Fig. 7. Sensitivity of VHL−/− ccRCC to EZH1 loss is driven by HIF-dependent induction of H3K27 demethylases.
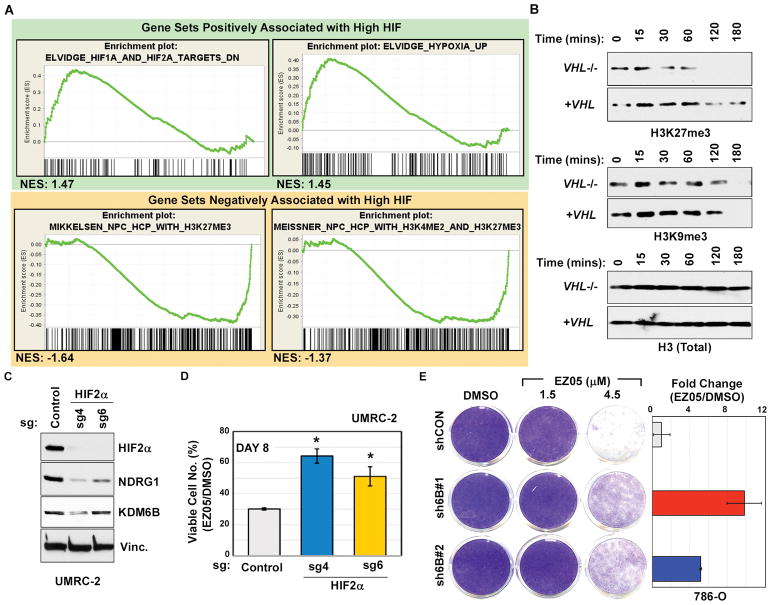
(A) GSEA of RNA-Seq data comparing gene expression in 786-O empty vector control cells versus 786-O cells in which ARNT1 was inactivated with either shRNA (3819, used in Fig. 6, E and F) or sgRNA (as in Fig. 6, G and H). Gene sets scoring with a FDR<0.25 and nominal p-value<0.05 were considered significant. NES indicates Normalized Enrichment Score.
(B) Immunoblots of purified histones incubated for the indicated time periods with lysates from UMRC-2 cells infected to produce wild-type pVHL (+VHL) or an empty lentivirus (VHL−/−).
(C and D) Immunoblots (C) and percent viable cells relative to DMSO-treated control (D) of UMRC-2 cells transduced to express sgRNAs targeting HIF2α (sgH2a4 and sgH2a6) or a non-targeting control. In (D), cells were treated with 3 μM JQ-EZ-05 or DMSO for 8 days. p-values were calculated using the Students t-test and p<0.05 was considered significant [*].
(E) (Left) Crystal violet staining of 786-O cells expressing either shRNA against KDM6B (JMJD3) (sh6B#1 and sh6B#2) or control shRNA (shCON) and treated with the indicated concentrations of JQ-EZ-05 for 8 days. (Right) Quantification of crystal violet stain in cells treated with 4.5 μM EZ-05 relative to cells treated with DMSO.