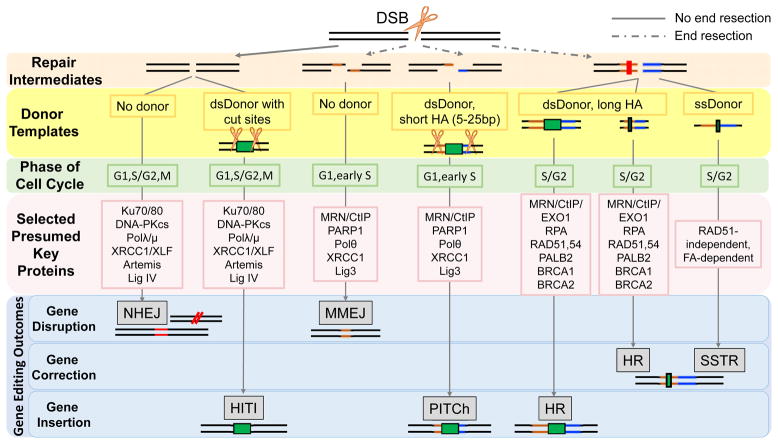
Figure 3. Summary of Gene Editing Pathways.
Double stranded break (DSB) is induced by a targeted nuclease (represented by scissors). DSB ends may or may not be resected (dashed or solid line, respectively),. The ultimate gene editing outcome (light blue boxes on the bottom) depends on several factors: the type of donor template provided (yellow box), the phase of cell cycle (light green box) and the presumed DNA repair proteins available (pink box). Gray boxes indicate the names of the repair mechanisms. It should be noted that the figure illustrates the common pathways described to date, however modification of DNA repair pathways and their utilization for gene editing purposes is an area of active research. (From left to right). A DSB with no end resection and no donor available is likely to result in insertions and deletions (indels) and lead to gene disruption via the non-homologous end joining (NHEJ) pathway. NHEJ may occur in any phase of cell cycle. Exogenously providing a double-stranded donor (dsDonor), which contains nuclease cut sites (scissors) around the gene of interest (green rectangle), may result in homology-independent targeted integration (HITI). The presence of microhomology on opposite strands of DNA around the cut site may result in gene disruption via the microhomology-mediated end joining (MMEJ) pathway. A recently reported method of gene integration, termed precise integration into target chromosome (PITCh), utilizes MMEJ machinery to integrate a gene of interest, which is provided by dsDonor with short homology arms (HA) to the DNA (HA are highlighted in orange and blue). The three pathways on the right are generally only active in S/G2 phases of cell cycle and may be used to correct a single nucleotide mutation in the DNA (represented by a red line). Exogenously providing a dsDonor with long homology arms may lead to either gene integration or gene correction via homologous recombination (HR) mechanism, depending on the length of the donor template. A new type of repair mechanism for gene correction was recently described, termed single stranded template repair (SSTR). Although resulting in the same outcome as HR-mediated gene correction, SSTR is presumed to utilize the Fanconi Anemia (FA) pathway and be RAD-51 independent.