(
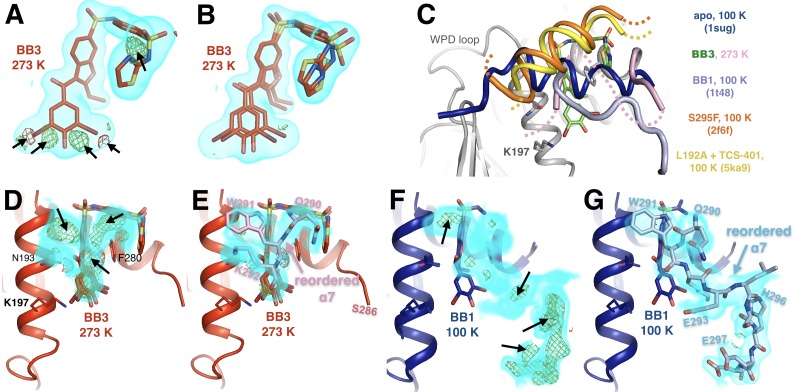
A) At 273 K, despite a good fit to 2Fo-Fc electron density, shown contoured at 0.75 σ (cyan volume), the allosteric inhibitor BB3 cannot be sufficiently modeled with a single conformer, as evidenced by +3.5 σ (green mesh) and −3.5 σ (red mesh) Fo-Fc difference electron density. These features are absent from the 100 K structure with BB3 (1t4j). (
B) These unexplained difference features disappear when a second alternate conformer is added with a translation at the ‘bottom’ of the molecule (from this viewing angle) and dihedral-angle changes at the ‘top right’. (
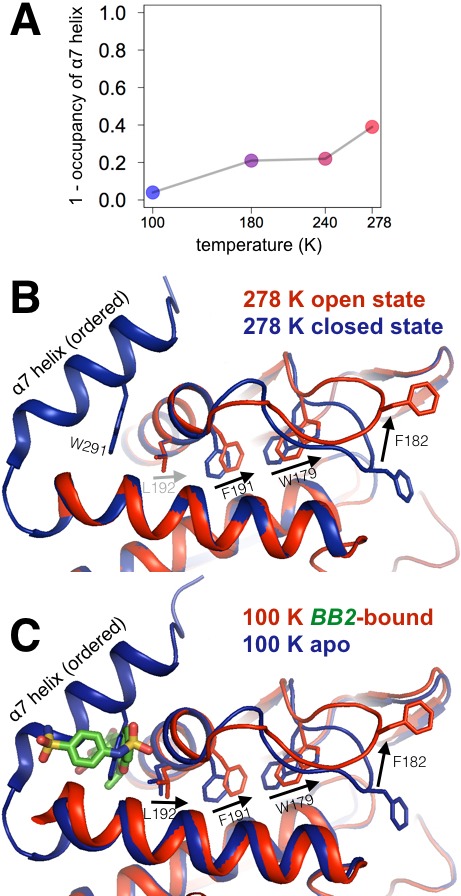
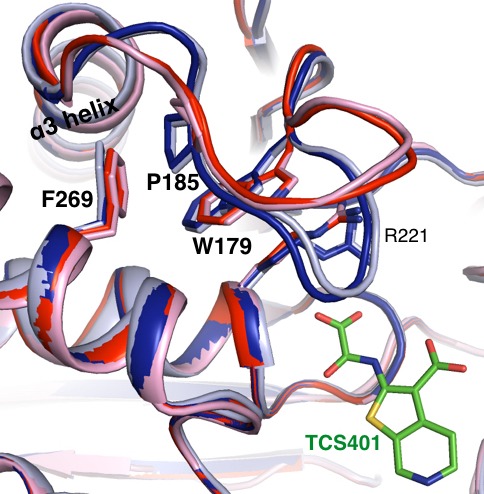
C) The disordered α7 helix is reordered above the BB binding site in various structures: 273 K with BB3 (pink; BB3 molecule in green), 100 K with BB1 (pale blue, PDB ID 1t48), 100 K with the S295F mutation (orange, PDB ID 2f6f), and 100 K with the L192A mutation and the TCS-401 active-site inhibitor (yellow, PDB ID 5ka9). The normal ordered α7 conformation (blue, PDB ID 1sug) is shown for reference. See
Figure 7A for another example of a reordered α7 conformation. (
D–G) Evidence for some of the conformations in C. (
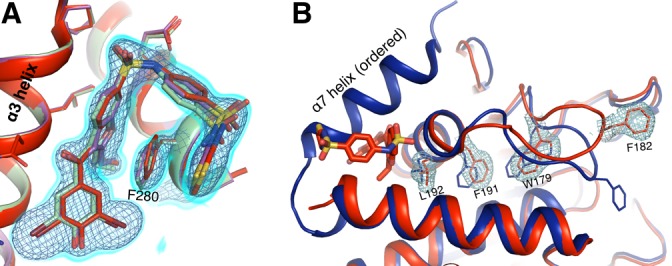
D) At 273 K, 2Fo-Fc electron density contoured at 1.0 σ (cyan volume) and Fo-Fc difference electron density contoured at +3.5 σ (green mesh) and −3.5 σ (red mesh) suggest something remains unmodeled above the bound BB3 molecule. (
E) Modeling a reordered portion of the disordered α7 helix, including Trp291 (pink), fits the 2Fo-Fc density, removes the positive Fo-Fc peaks, and has reasonable interactions with nearby sidechains on α3 (left). (
F–G) At 100 K, the electron density (shown at the same contour levels) suggests a different reordered conformation of α7 above BB1 (PDB ID 1t48). Both reordered α7 conformations (
D–E) vs. (
F–G) place residues 290–292, including Trp291, in the same place. However, the C-terminal portion of the reordered conformation with BB1 at 100 K, including His296 (right), is sterically incompatible with the N-terminal portion of the α6-α7 junction with BB3 at 278 K, including Ser286. Neither conformation is compatible with the electron density for the other, suggesting that differences in temperature and/or inhibitor dictate different α7 conformations. The viewing orientation in
C–G) is as in
Figure 1B (‘back side’ of PTP1B), except zoomed in on the BB site (labeled in
Figure 1B).