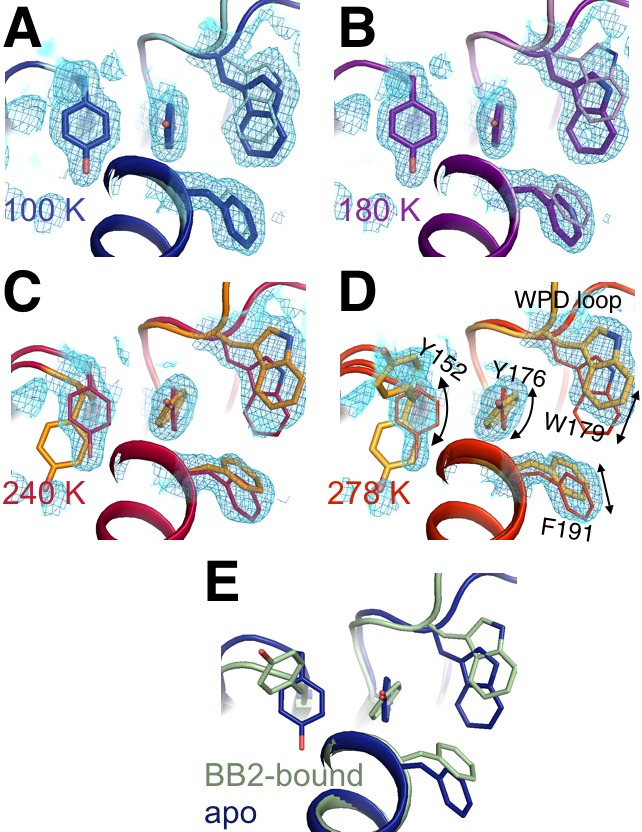
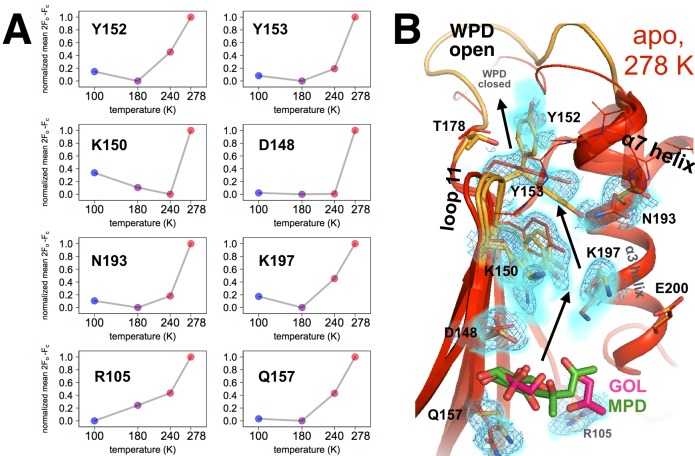
Figure 5. Coupled conformational heterogeneity leads to the allosteric 197 site.
(A) Several residues distinct from both the active site and a previously characterized allosteric network each have minor alternative conformations that become more populated with temperature. This is quantified by the sum of 2Fo-Fc electron density values for the centers of atoms that are unique to the minor state (defined as being at least 1.0 Å away from any atoms in the major state), normalized across temperatures from 0 to 1 for each residue. (B) These residues colocalize to a region of the protein surrounded by loop 11 (top-left), the quasi-ordered α7 helix (top-right), and the α3 helix (right), including the eponymous K197. 2Fo-Fc electron density contoured at 0.6 σ (cyan volume) and at 0.8 σ (blue mesh) justify multiple conformations for these residues in our 278 K apo model, as quantified in (A). The alternative conformations of these residues appear to interact with one another and thus may be allosterically coupled. Ordered crystallization mother liquor or cryoprotectant molecules (glycerols in pink, from the PDB and our structures, or MPD molecules in green, from the PDB) can be present at the terminus of this allosteric pathway, suggesting it may be amenable to binding other small molecules. The viewing orientation in B) is as in Figure 1B (‘back side’ of PTP1B), except zoomed in on the 197 site (labeled in Figure 1B).
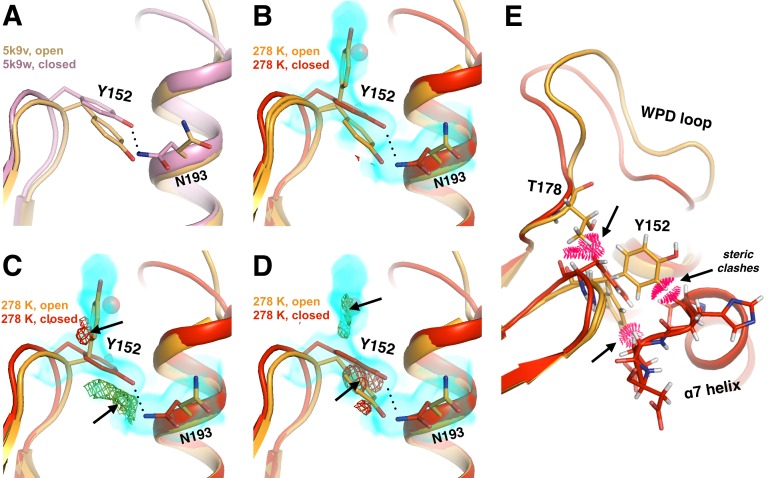
Figure 5—figure supplement 1. Alternative conformations in apo PTP1B recapitulate and expand upon reported coupling between loop 11 and α3.
Figure 5—figure supplement 2. The allosteric 197 site has local sequence differences in related PTPs.
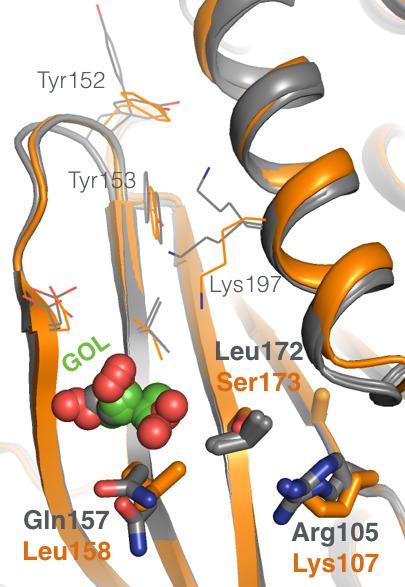
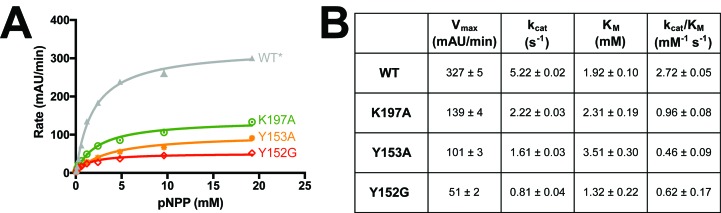
Figure 5—figure supplement 3. Mutations along the 197 site’s allosteric pathway reduce enzyme activity.
Figure 5—figure supplement 4. Flexible aromatic residues complete an allosteric circuit.