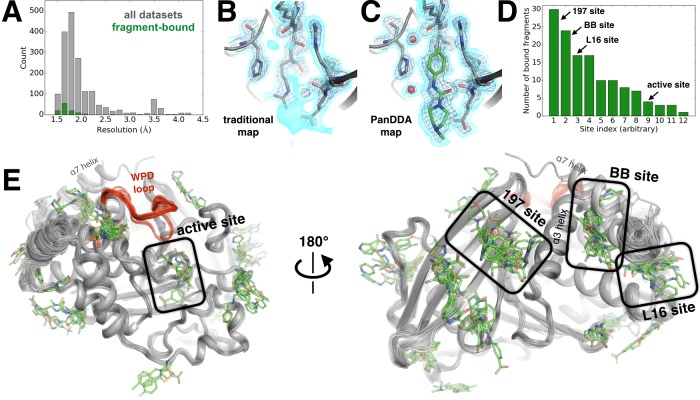
Figure 6. Electron-density background subtraction reveals small-molecule fragments at allosteric sites in PTP1B.
(A) Histogram of X-ray resolution for 1774 structures of PTP1B soaked with small-molecule fragments (gray) vs. the 110 structures from that set with small-molecule fragments bound to PTP1B (green). (B) For one example fragment, a traditional 2Fo-Fc map contoured at 1.25 σ (cyan volume) and at 3.5 σ (blue mesh) provides no clear evidence for a bound fragment. (C) By contrast, a background-subtracted PanDDA event map (85% background subtraction in this case) contoured at the same levels clearly reveals the precise pose of the bound fragment, plus additional ordered water molecules that accompany it (red spheres). (D) PanDDA analysis and manual inspection reveal 110 fragment-bound structures of PTP1B, with bound fragments clustered into 12 non-overlapping binding sites. Some structures contain multiple bound copies of the same fragment. Several sites of interest are labeled. (E) Overview of bound fragments across the PTP1B surface. Left: front of protein, facing active site (WPD loop open and closed conformations in red). Right: back of protein, facing several fragment-binding hotspots: the 197 site, BB site, and L16 site. The viewing orientation in E) (left) is as in Figure 1A (‘front side’ of PTP1B). The viewing orientation in E) (right) is as in Figure 1B (‘back side’ of PTP1B).