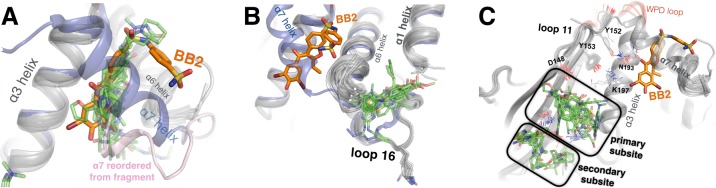
Figure 7. Fragments cluster at three binding hotspots distal from the active site.
(A) Twenty-four fragments (green) bind to the same site and in similar poses as the BB2 inhibitor (orange, PDB ID 1t49), and similarly displace the α7 helix (foreground, transparent blue, PDB ID 1sug). BB2 is also shown in the following panels to emphasize that its binding site is distinct from the other fragment-binding hotspots. One structure with a fragment bound in this site features a reordered conformation of the α7 helix (pink). (B) Seventeen fragments bind to the L16 site, where they may modulate the conformations of loop 16, the α6 helix, and the protein’s N-terminus on the α1 helix. (C) Thirty fragments bind to the 197 site in one primary subsite contacting K197, or a distinct secondary subsite nearby. The viewing orientation in (A) is as in Figure 1B (‘back side’ of PTP1B), except zoomed in on the BB site (labeled in Figure 1B). The viewing orientation in (B) is also as in Figure 1B, except looking left from the right of that image and zoomed in on the L16 and BB site site. The viewing orientation in A) is as also in Figure 1B, except zoomed in on the 197 site and BB site (labeled in Figure 1B). See also Figure 6E (right) for orientation.
Figure 7—figure supplement 1. Fragments overlap with the BB allosteric inhibitor scaffold and suggest possible improvements.
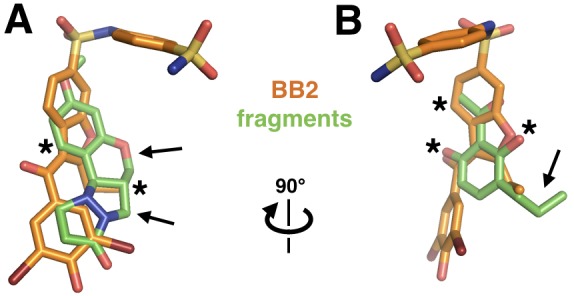
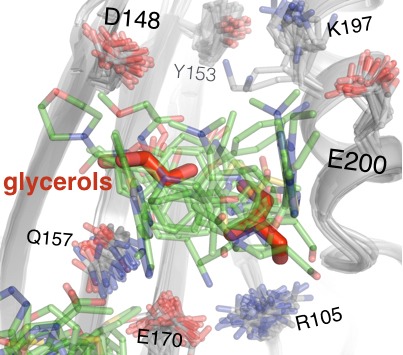
Figure 7—figure supplement 2. Fragments in the 197 site overlay with glycerols from multitemperature structures.