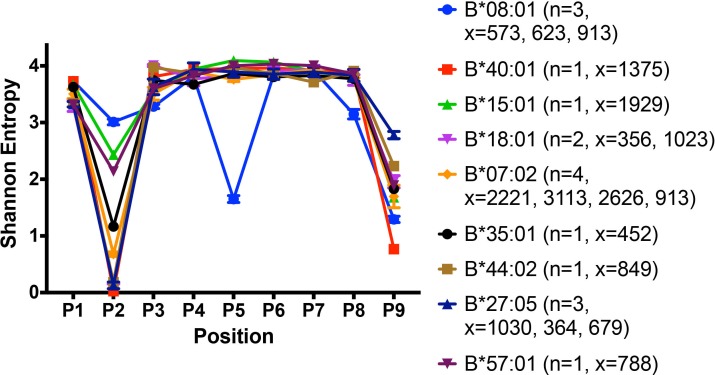
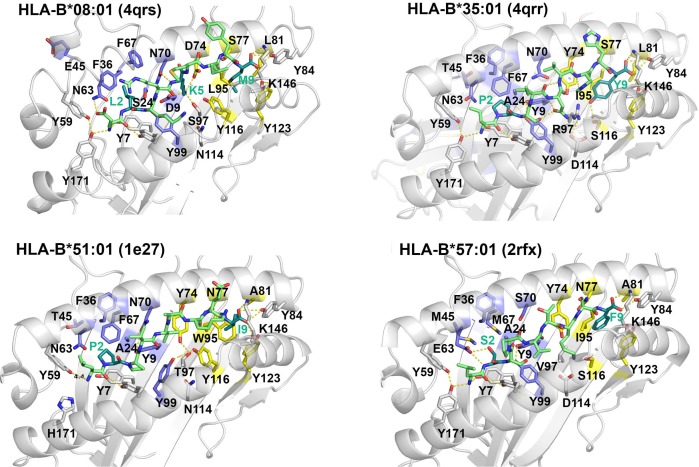
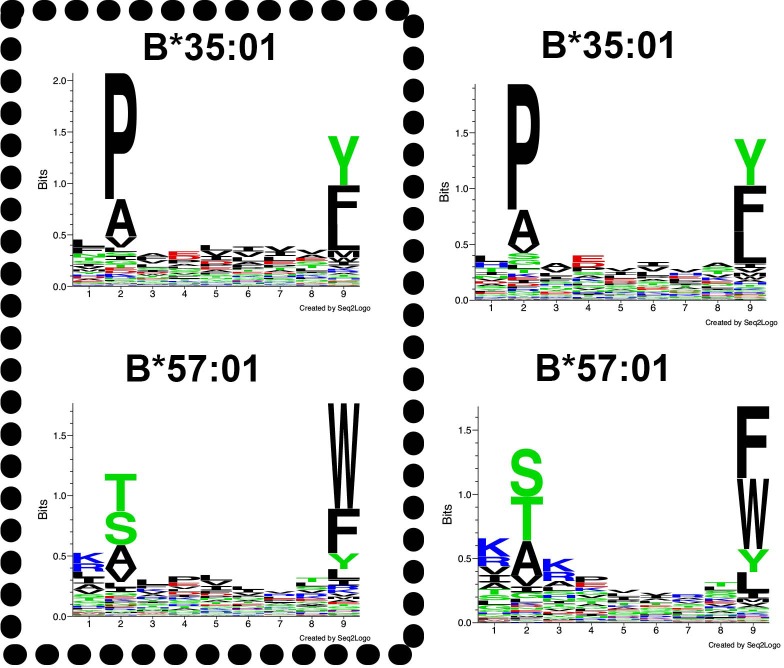
Figure 8. Peptidome and peptide-binding characteristics of HLA-B.
Shannon entropy plots of B lymphoblastic cell-derived 9-mer peptides for the indicated allotypes, based on mass spectrometry datasets obtained from (Pearson et al., 2016). Plots are based on 9-mer peptides assigned to each allele based on IEDB predictions (iedb.org). n values represent the total number of independent datasets used for the plots. x values represent the number of peptides in the independent datasets used for the plots. This figure has two supplementary figures.