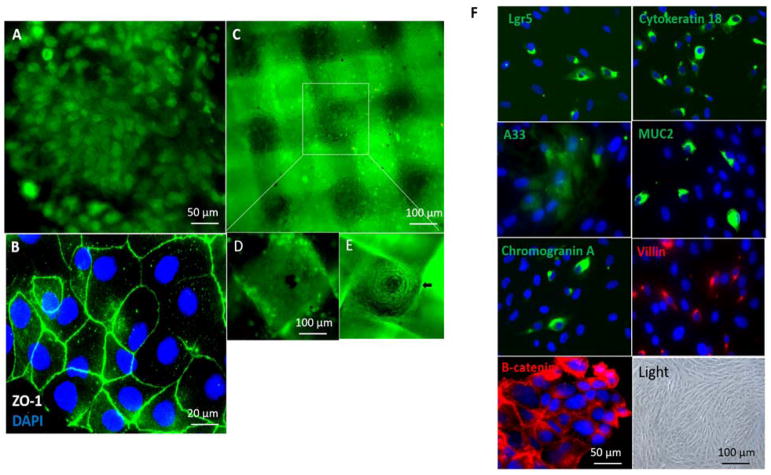
Figure 3. Co-culture of hIECs and C3A cells on the GI-Liver module.
The devices were cultured with a 1:1 mixture of medium from each of the 2 cell types with addition of 3% serum. After co-culture for 14 days, the devices were disassembled and stained with Invitrogen’s Live/Dead Stain or stained for tight junctions (ZO-1, green) and the nuclei counterstained with DAPI (blue). Cells were imaged under a fluorescent microscope. Scale bars are present on the micrographs. A. hIECs formed a monolayer in the GI-chamber and was stained with 2 μM calcein AM for live/dead cell assays. B. hIECs were stained with ZO-1 and DAPI. C. HepG2 C3A cells were plated on 3D construct and stained with 2 μM calcein AM. D. and E. The apical and basolateral surfaces of the 3D liver culture on a scaffold in which the C3A cells formed micro-liver lobe structure. F. Characterization of the hIECs cultured in the GI-chamber: cells expressed the stem cell marker Lgr5, epithelial markers (cytokeratin 18, colon epithelial cell-specific marker A33, β-Catenin in WNT signaling) and differentiation markers (villin (absorptive cells), mucin 2 (goblet cells) and chromogranin A (enteroendocrine cells)), detected by immunostaining. Nuclei were counter-stained (DAPI, blue). A bright field micrograph was included.