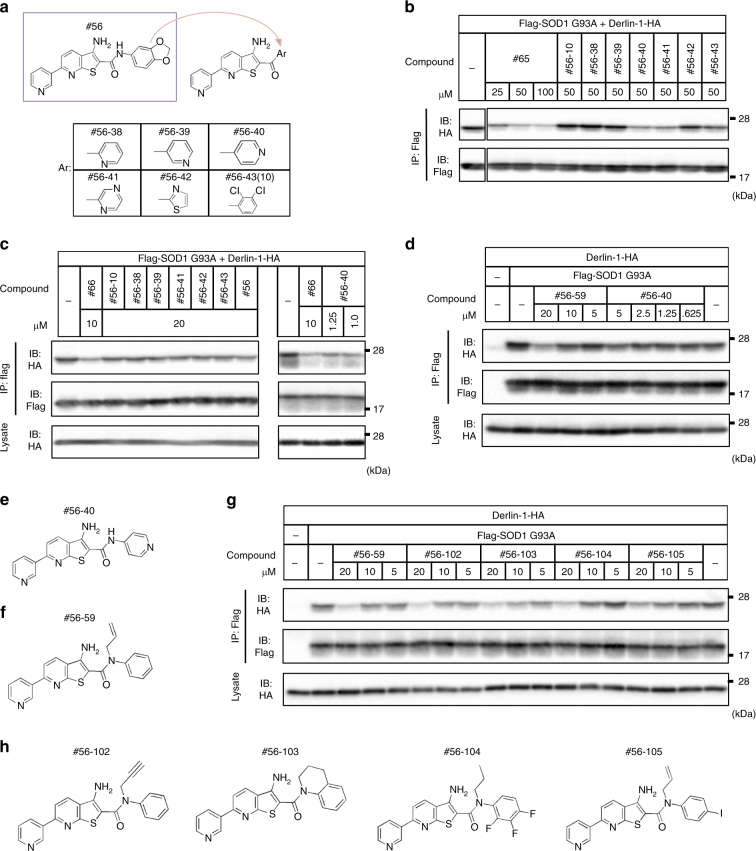
Fig. 3.
Identification of cell-permeable compounds. a Chemical structure of newly synthesized #56 analogs. #56-43 is the resynthesized #56-10. b Inhibition of SOD1G93A-Derlin-1 interaction by new #56 analogs in in vitro IP assay. The SOD1G93A-Derlin-1 complex purified from lysates transfected with Flag-SOD1G93A and Derlin-1-HA was incubated with the indicated compounds for 16 h and analyzed by IP-IB with the indicated antibodies. c, d Inhibition of SOD1G93A-Derlin-1 interaction by #56 analogs in a cell-based IP assay. HEK293A cells transfected with indicated plasmids were treated with the indicated compounds for 24 h, and lysates were analyzed by IP-IB with the indicated antibodies. c Inhibition of SOD1G93A-Derlin-1 interaction by the same compounds as in the in vitro assay b in a cell-based assay. Compound #66 is used as a positive control of a cell-permeable inhibitor, although the resynthesized #66 does not show any activities. d Two cell-permeable SOD1G93A-Derlin-1 interaction inhibitors in a cell-based IP assay. e, f Chemical structure of #56-40 and #56-59. g Inhibitory activities of other compounds containing tertiary amide groups, #56-102-105, in the cell-based IP assay as in c, d. h Chemical structure of #56-102, 103, 104, and 105