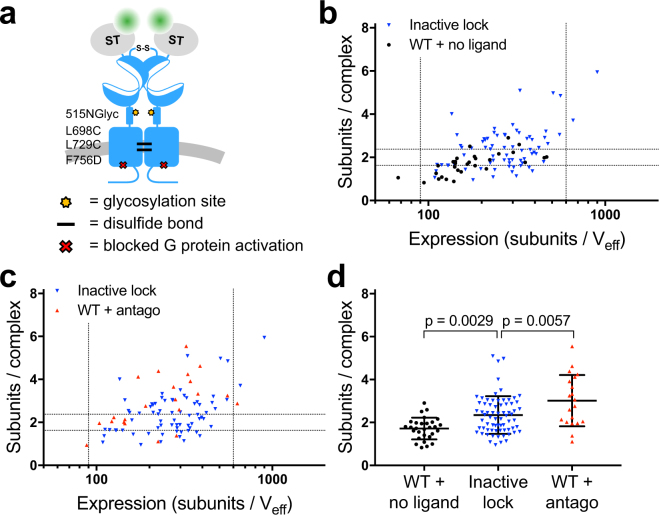
Figure 6.
Locking the inactive conformation of mGlu2 allows its oligomerization in absence of ligand. (a) Schematic drawing of the mutations introduced to lock the inactive conformation of the mGlu2 receptor. An N-glycosylation site was introduced at residue 515 (515NGlyc) in the CRD to inhibit contact between the CRDs of the dimer and cysteines were introduced in TM4 and TM5 (L698C and L729C) to cross-link the inactive interface of the 7TM dimer. In addition a mutation in intracellular loop 3 that inhibits G protein activation (F756D) was introduced. (b,c) Stoichiometry as a function of the expression for mGlu2 locked in the inactive conformation compared with wild type receptor incubated with CuP and subsequently imaged in the absence of ligand (b) or bound to antagonist (c). (d) The number of subunits per complex for neurons with expression in the 90–600 subunits/Veff range. In this range the mean receptor stoichiometry is 2.3 ± 0.9 subunits/complex for mGlu2 locked in the inactive conformation (70 cells), 1.7 ± 0.5 subunits/complex for wild type mGlu2 in the absence of ligand (29 cells), and 3.0 ± 1.2 subunits/complex for wild type mGlu2 bound to antagonist (20 cells). The mean stoichiometry of mGlu2 locked in the inactive conformation is significantly higher than wild type mGlu2 in the absence of ligand (p = 0.0029, one-way ANOVA, Bonferroni’s multiple comparisons test). Note that these apparent stoichiometry values are lower in the presence than in the absence of CuP (Supplementary Fig. S8). Errors and error bars are SD. Data are from 3–6 (mGlu2) or 10 (inactive lock) independent experiments.