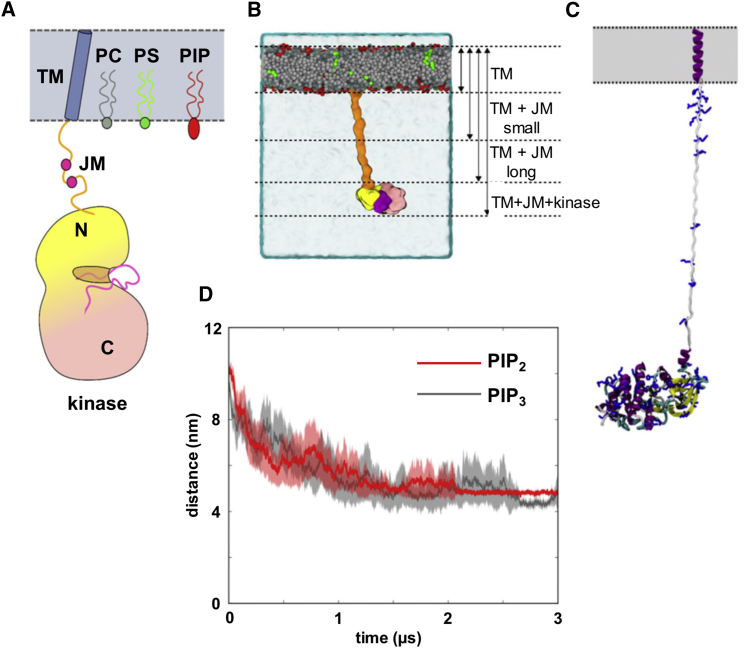
Figure 1.
The Juxtamembrane and Kinase Domains of EphA2 at the Membrane
(A) Schematic representation of the first (i.e., N-terminal part) of the cytosolic domains of EphA2, showing the transmembrane (TM; blue) helix domain followed by a juxtamembrane (JM; orange) segment, and the kinase domain (N-terminal lobe in yellow, the C-terminal lobe in pink, and the activation loop in pink). The lipid bilayer is shown in gray with the three classes of lipid included in our simulations: phosphatidylcholine (PC; gray), phosphatidyl serine (PS; green) and phosphatidyl inositol phosphates (PIP; in red). The two mauve circles in the JM region depict the conserved Tyr motif (Y588 and Y594 for EphA2) that can be phosphorylated.
(B) Coarse-grained model of the EphA2 JM + kinase domains tethered at a membrane by the TM domain (hidden by lipids). In the starting configuration, we modeled the JM segment as extended but flexible. Upon simulation the JM domain collapsed onto the bilayer surface drawing the kinase domain toward the membrane (see Video S1). Simulations (see Table 1 for details) were performed for systems with just the TM domain with one or two extensions of the JM domain in addition to the TM + JM + kinase system depicted.
(C) Starting tethered model in atomistic resolution showing the positively charged residues in the JM and kinase domains.
(D) Evolution of the average distance between the centers of mass of membrane and the untethered kinase domain for simulations (see Table 1) in which the bilayer contained PIP2 (red) or PIP3 (gray). A distance of 4–5 nm indicates a stable interaction between the kinase and the membrane. In each case the bold line shows mean distance for each set of ten simulations and the transparent background the SEM. See also Figure S1 for further details of these simulations.