Figure 2.
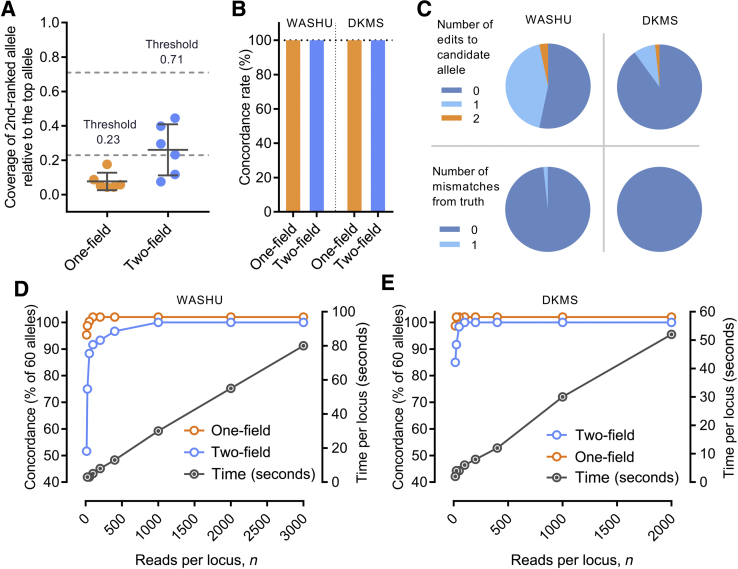
Human leukocyte antigen (HLA) typing accuracy and read downsampling. A: The ratios of the coverage of second-ranked allele to that of the top-ranked allele were plotted for the six homozygous samples in the Washington University–Training (WASHU-T) data set at one-field and twp-field levels, respectively. Dashed lines are heterozygosity thresholds, 0.23 and 0.71, which are the means of the ratios plus 3 SDs at one-field and two-field levels, respectively. B: Concordance of HLA typing results for the Washington University (WASHU) and German Marrow Donor Program (DKMS) data sets at one-field and two-field resolutions. A horizontal reference line at the concordance rate of 100% is shown (dotted line). C: Percentage of candidate alleles with zero, one, or two edits (top panel) and percentage of consensus sequences with zero or one mismatch from the reference sequence of true alleles (bottom panel) in the WASHU and DKMS data sets. D and E: Effect of downsampling of the WASHU (D) and DKMS (E) data sets on the concordance rates. Concordance at one-field and twp-field resolutions and the computation time were plotted against the numbers of reads per locus. Data are expressed as means ± SD (A).