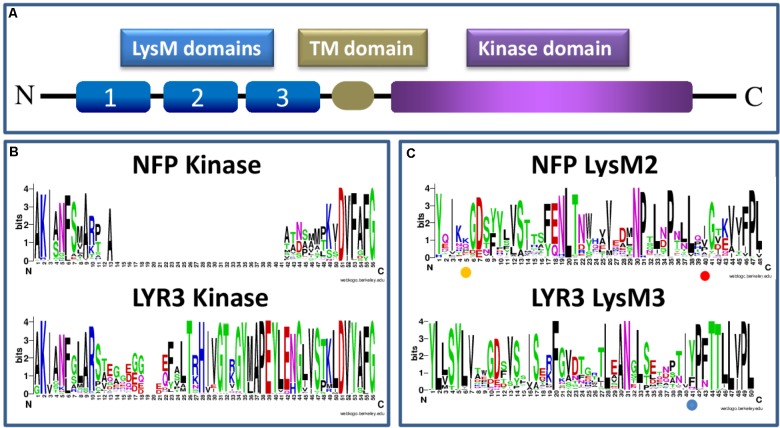
FIGURE 1.
Schematic presentation of a LysM-RLK protein structure (A) and sequence logos generated using Clustal W (Larkin et al., 2007) and WebLogo software (Schneider and Stephens, 1990; Crooks et al., 2004), and approximately 60 Angiosperm protein sequences each of NFP or LYR3 (B,C) (Supplementary Table S1 and Supplementary Data Sheet S1). The sequence logos show the extent of sequence conservation within (B) part of the intracellular domain that contains the activation loop in LYR3 proteins but not in NFP proteins; and (C) individual LysM domains (LysM2 for NFP proteins and LysM3 for LYR3 proteins). In (B) the conserved “F” at position 6 of the sequence logos corresponds to the “F” in the “DFG” motif of active kinases. The amino acid positions highlighted in (C) correspond to L118 in LjNFR5 LysM2; L154 in MtNFP LysM2 and Y228 in MtLYR3 LysM3, as discussed in the text. The LysM sequence logos also show that each LysM domain is a combination of highly conserved and highly variable residues, many of which are probably involved in tertiary structure and ligand binding specificity, respectively. Sequence logos of all the 3 LysM domains of NFP and LYR3 are shown in Supplementary Figures S1, S2.