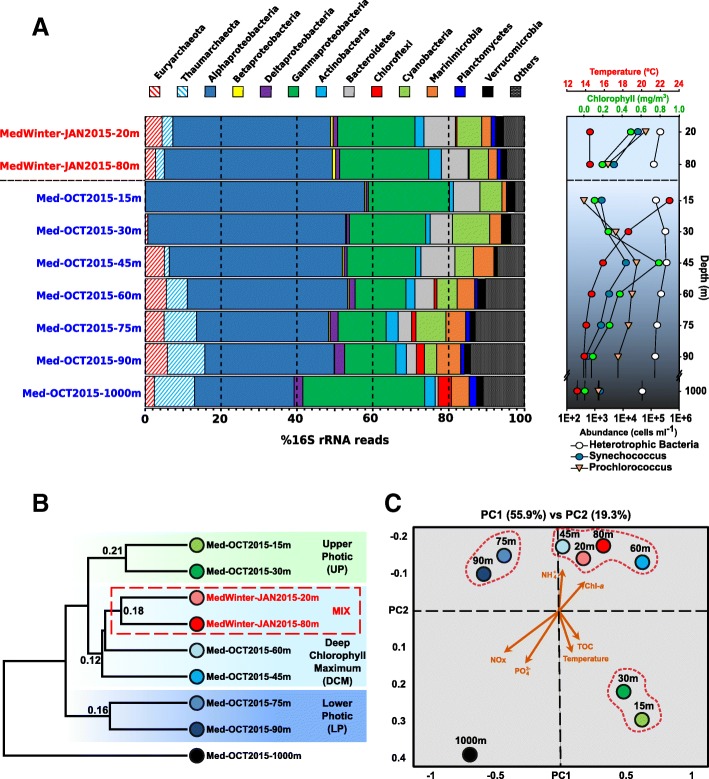
Fig. 1.
a Phylum-level composition based on 16S rRNA gene fragments (raw reads) of the different metagenomes. The phylum Proteobacteria was divided into its class-level classification. Only those groups with abundance values larger than 1% in any of the metagenomes are shown. Sample names in blue and red correspond to those collected in stratified and mixed water columns, respectively. On the right panel, a vertical profile of the temperature, chlorophyll-a, and abundance of heterotrophic bacteria, Synechococcus and Prochlorococcus cells is shown. The horizontal dashed line separates the mixed samples from the stratitied samples. b Dendrogram of metagenomic dataset raw sequence similarity. Samples highlighted with a red dotted line were collected in winter. Numbers at each node indicate similarity (1 would be 100% similarity) among the samples using raw reads. c Canonical correspondence analysis (CCA) between physicochemical parameters and read annotation similarity. Chl-a, chlorophyll-a; TOC, total organic carbon