Abstract
A hallmark of metazoan evolution is the emergence of genomic mechanisms that implement cell type-specific functions. However, the evolution of metazoan cell types and their underlying gene regulatory programs remain largely uncharacterized. Here, we use whole-organism single-cell RNA-seq to map cell type-specific transcription in Porifera (sponges), Ctenophora (comb jellies) and Placozoa species. We describe the repertoires of cell types in these non-bilaterian animals, uncovering diverse instances of previously unknown molecular signatures, such as multiple types of peptidergic cells in Placozoa. Analysis of the regulatory programs of these cell types reveal variable levels of complexity. In placozoans and poriferans, sequence motifs in the promoters are predictive of cell type-specific programs. In contrast, the generation of a higher diversity of cell types in ctenophores is associated to lower specificity of promoter sequences and to the existence of distal regulatory elements. Our findings demonstrate that metazoan cell types can be defined by networks of TFs and proximal promoters, and indicate that further genome regulatory complexity may be required for more diverse cell type repertoires.
The origin of animal multicellularity was linked to the spatial co-existence of cell types with distinct roles1,2. Cell type specialization is achieved through asymmetric access to genomic information, which is interpreted in a cell-specific fashion through mechanisms of transcriptional gene regulation. However, it remains unclear how elaborate genome regulation relates to cell type diversity. Poorly characterized, early-branching metazoans represent an opportunity to explore these questions by studying how cell type-specific genome regulation is implemented in species with (presumed) intermediate to low organismal complexity. Sponges, comb jellies and placozoans are, together with the remaining animals (Planulozoa), phylogenetically the earliest-branching animal lineages3–6 (Fig. 1). These organisms possess characteristic body plans and have been traditionally considered to contain low numbers of cell types7, although our current understanding of this diversity of cell behaviors remains very limited. Moreover, these three lineages have diverged for over 650Ma8, which has resulted in extremely different and specialized morphologies, life strategies, and body plan organization9. Ctenophores are marine predators (mostly pelagic), they have tissue-level organization, and they develop a nervous system of uncertain homology with their bilaterian counterparts10–12. In contrast, sponges are sessile filter-feeders that live both in marine and freshwater environments and that seem to have no or very rudimentary specialized tissues13. Finally, placozoans are tiny benthic marine animals with a bodyplan organization that is composed out of two cell layers, they possess ciliary-based locomotion, and they feed on algae using external digestion14.
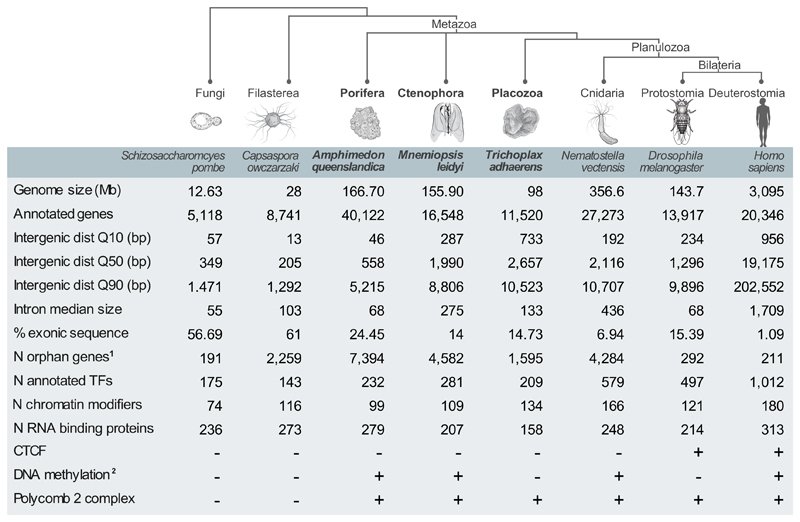
Figure 1. Comparison of genomic features of early metazoans and phylogenetically-related species.
Lineages/species sampled in this study are highlighted in bold. 1Number of orphan genes based on Ensembl (second value), except for Capsaspora owczarzaki (based on48). 2Presence/absence of DNA methylation in species without methylation data based on presence/absence of Dnmt1/3 orthologues.
Sponges, ctenophores and placozoans also vary considerably in their overall genome size, median intergenic space, and repertoire of potential transcriptional and post-transcriptional regulators (Fig. 1). The genome of the sponge Amphimedon queenslandica measures 166mb, and its annotation suggests a relatively compact gene arrangement with very short (0.6kb) intergenic regions15,16. In comparison, similar genome size (156mb) but longer (2kb) intergenic regions are found in the ctenophore Mnemiopsis leidyi17. In the case of the placozoan Trichoplax adhaerens, a smaller genome (98mb) but longer intergenic regions (2.7kb) are reported18. Annotation and comparison of the predicted proteome in these non-bilaterian species uncovered an extensive suite of gene families shared across Metazoa15,17–19, suggesting the existence of ancient regulatory mechanisms for orchestrating cell type specification and maintenance. For example, sponge, ctenophore and placozoan genomes encode for substantial repertoires of transcription factors (209-232) and chromatin modifiers/remodelers (99-134), representing intermediate diversity compared to unicellular species and to other metazoans (e.g. cnidarians or bilaterians) (Fig. 1). However, comparative analysis of genomic regulatory programs in non-model organisms is confounded by the scarcity of direct molecular data on cell states and genome regulation. Whole-organism single cell RNAseq20,21 opens an opportunity to start closing this gap, by performing extensive sampling of transcriptional programs and characterizing cell type repertoires in diverse metazoan lineages. Here, we generate transcriptional maps at single cell resolution for A.queenslandica, M.leidyi and T.adhaerens. These maps, in combination with chromatin data and sequence analysis, allow us to survey the cell type diversity and to compare the genomic regulatory programs in these non-bilaterian animal lineages.
Results
An atlas of Amphimedon queenslandica adult and larval cell types
In order to study sponge cell type diversity, we collected adult and larval specimens from A.queenslandica. We processed fresh cells using the MARS-seq protocol with small adaptations22 (see Methods), profiling in total 4,992 adult and 3,840 larval A.queenslandica cells (Supplementary Fig. 1, Table S1). Whole-organism single cell analysis involves processing of cells with highly heterogeneous RNA content, given the expected differences in size and/or transcriptional activity between distinct cell types (Supplementary Fig. 1a-b). To maximize the sensitivity of our assay, we retained for subsequent analysis all sampled cells with at least 100 unique molecule identifiers (UMI). Applying the MetaCell framework (Appendix S1), we found over 300 marker genes in each stage, which showed high degree of intra-population transcriptional variance (Supplementary Fig. 1c). Using this approach, even cells with overall low UMI counts were characterized by a sufficient number of marker genes (Supplementary Fig. 1d). This allowed us to robustly group 81-94% of our single cells into transcriptionally coherent clusters (that we call metacells, see Methods and Appendix S1) (Supplementary Fig. 1e-f), and to apply a bootstrap approach to support these metacells (see Methods) (Supplementary Fig. 1g). Moreover, we associated each of the derived metacells with a set of differentially expressed genes (Table S2 and S3) and used the functional annotation of these gene sets to annotate at least some of the metacells.
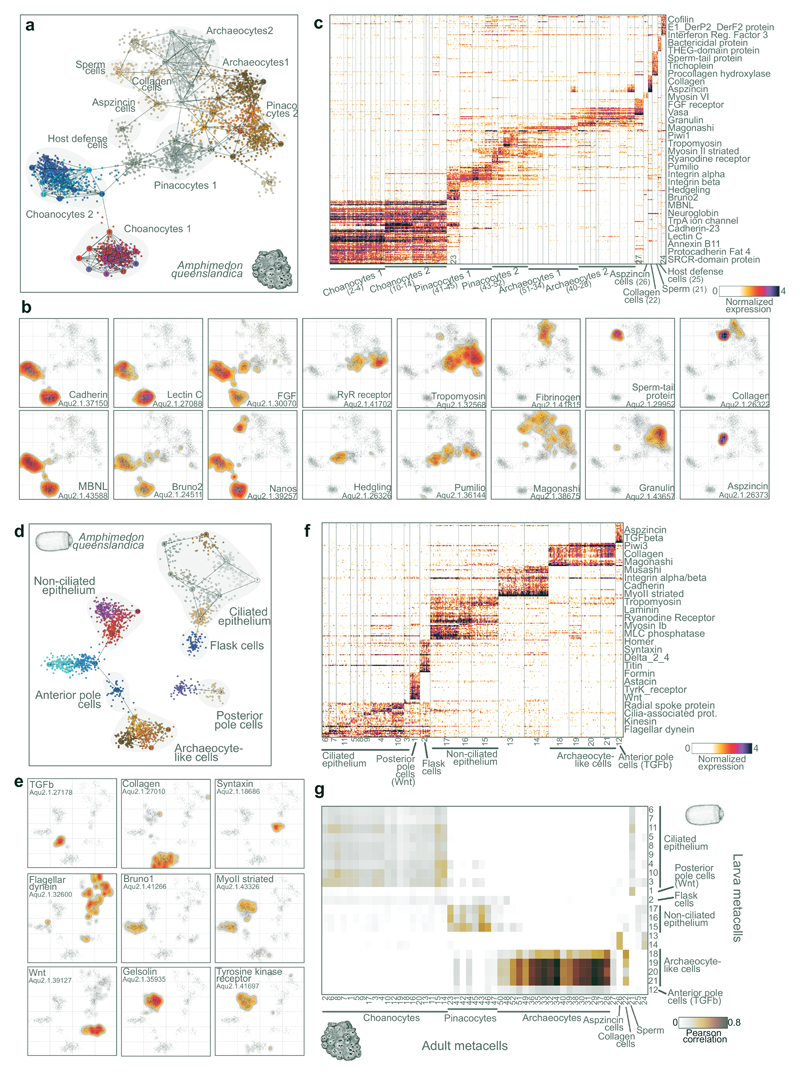
The power of whole-organism single cell RNAseq analysis to characterize cell types is demonstrated by visualizing A.queenslandica adult metacells (Fig. 2a), key marker genes projected in 2D (Fig. 1b), and a heatmap showing distribution of marker genes at single cell resolution (Fig. 2c). The sponge transcriptional landscape is dominated by large groups of choanocytes, pinacocytes and archaeocytes13. Even though these groups can be further subdivided into subclasses, their annotation into broad types is supported by common transcriptional signatures of key genes. Choanocytes are autonomous filter-feeding cells with a unique morphology, characterized by a flagellum surrounded with a microvilli collar23. Our data shows that A.queenslandica choanocytes express RNA-binding proteins like MBNL, Bruno2 and Nanos; multiple proteins of the flagellar apparatus; and annexins24 (Fig. 2b, Supplementary Fig. 2b,h). They also specifically express multiple adhesion proteins including cadherins and C-type lectins (Fig. 2b). Interestingly, not only choanocytes, but also other cell types we identified express unique combinations of adhesion proteins, for example distinct integrin alpha/beta paralog pairs (Supplementary Fig. 2a). These cell type-specific adhesion molecules, especially those like cadherins and immunoglobulins that mediate homophilic interactions, are likely to be important in the spatial sorting of cell types and general sponge body plan organization. Finally, based on their expression, we can define two broad types of choanocytes (Fig. 2a) showing differences not only in their repertoire of effector genes but also in the expression of transcription factors (see Fig. 5d).
Figure 2. Amphimedon queenslandica adult and larva cell type atlases.
a, 2D projection of A.queenslandica adult metacells and single cells. Cell clusters with known/hypothesized identity are annotated and highlighted in grey. b, Gene expression distribution on 2D projected A.queenslandica adult cells for selected gene markers. Cells 2D projection is the same as in a. c, Normalized gene expression across 3,870 A.queenslandica adult single cells (columns), sorted by cell cluster. For each cluster, the top 25 genes sorted by fold change ver sus the other metacells were selected for visualization (with a FC threshold>=2). d, 2D projection of A.queenslandica larval metacells and single cells. e, Gene expression distribution on 2D projected A.queenslandica larval cells for selected gene markers. Cells 2D projection is the same as in d. f, Normalized gene expression across 1,932 A.queenslandica larva single cells (columns), sorted by cell cluster. Genes selected as in c. g, Comparison of adult versus larval cell clusters. The heatmap shows the correlation values between metacells based on highly variable genes (FC>2 in at least 1 adult and 1 larval metacell). Notice the strong association between adult archaeocytes and a group of larval cells, suggesting the re-usage of this specific cell type program in two different post-embryonic stages. Color-coding of cells and metacells in a and d is arbitrary.
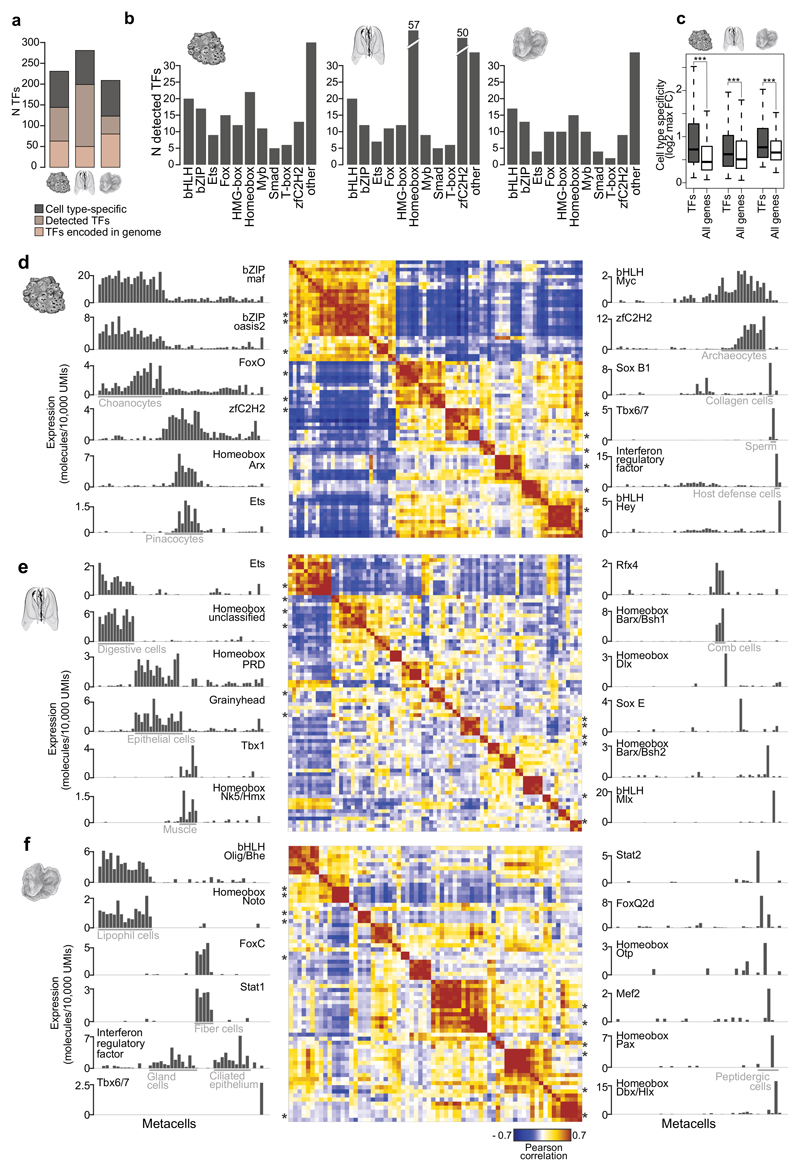
Figure 5. Transcription factor regulatory programs in A.queenslandica, M.leidyi and T.adhaerens.
a, Number of TFs encoded in the genome, detected in our scRNAseq analysis and showing cell type-specific expression in each species. b, Number of TFs belonging to different structural classes detected in each species. c, Cell type specificity of TFs compared to all genes, in each species. Cell type specificity of each gene is measured as the max fold-change enrichment of its expression in any metacell. *** p<<0.001; Wilcoxon rank-sum test. d, Heatmap (center) showing A.queenslandica TF-TF correlation based on expression profiles across metacells. Only TFs with >20 total molecules and FC>1.8 in at least one metacell are included. On both sides, barplots show the expression profile across metacells for representative TFs in each TF module. Asterisks indicate the position of the TFs shown in barplots in the heatmap (in the same descending order). e, f, Same as d for M.leidyi and T.adhaerens.
Another abundant group of cells we identified are pinacocytes (Fig. 2a,c). Pinacocytes are epidermal cells that cover the outer and inner surfaces of the sponge13. Our data shows that A.queenslandica pinacocytes specifically express Pumilo RNA-binding protein and multiple components of the actin contractility apparatus, including tropomyosin, calponin and striated-type myosin II (Fig 2b, Supplementary Fig. 2a). This suggests that A.queenslandica pinacoderm has some contractile properties, as also indicated by experiments in the demosponge Tethya wilhelma25. Interestingly, we also identify a cluster of cells that show intermediate transcriptional profiles between choanocytes and pinacocytes, expressing both choanocyte markers like FGF and Bruno2 and pinacocyte markers like Pumilio (Fig. 2b). In addition, these cells express specifically Hedgling (Fig. 2b), a cadherin with a N-terminal hedgehog domain26,27. This data suggest the existence of transcriptional states representing trans-differentiation intermediates between cell types, a process known to occur in multiple sponge species, including A.queenslandica13,28.
The last major sponge cell behavior that we identify correspond to archaeocytes, which are pluripotent amoeboid cells found in the sponge mesohyl (the gelatinous matrix that fills the sponge body)29. We find that these cells express specific extracellular matrix proteins (like fibrinogen), granulins, and large amounts of diverse RNA-binding proteins (like Magonashi) (Fig. 2b, Supplementary Fig. 2b-c). The extensive usage of cell type-specific RNA-binding proteins observed chiefly in archaeocytes, but also in other sponge cell types (Fig. 2b, Supplementary Fig. 2b), is in line with previous reports that suggest a pervasive role of this type of regulators in another sponge species: Ephydatia fluviatilis30. In addition to these abundant cell types, we detect in adult A.queenslandica remarkably distinct, yet much less abundant, cell types. These include sperm cells, defined by expression of TPRV ion channel, THEG, and other genes associated to sperm function (Fig. 2b, Supplementary Fig. 2d); collagen-producing cells (Fig. 1b); cells expressing multiple aspzincin protease paralogs (Fig. 2b, Supplementary Fig. 2e); and host defense cells producing anti-bacterial proteins (Supplementary Fig. 2f).
Unlike the other species included in this study but similar to many marine invertebrates31, A.queenslandica has a bi-phasic life cycle involving two dramatically different post-embryonic stages: adult and larva32. We therefore profiled single-cell transcriptomes in the lecitotrophic larva of A.queenslandica, in order to identify larval cell types and to compare them to those found in adult sponges. We profiled the transcriptomes of 3,840 larval single cells and identified metacells with specific expression signatures using the same strategy described for the adult (Fig. 2d-e, Table S2). This analysis revealed at least 7 different cell types in the larva (Fig. 2d-e). Based on published expression patterns for markers genes we could identify some of these cell types. These include ciliated epithelial cells that express ciliary markers (Fig. 2e), flask cells33, Wnt-expressing posterior pole cells34, and TGFβ-expressing anterior pole cells34. When comparing transcriptional signatures, larval cell types show remarkable differences with adult cell types. 4.8% of the genes expressed in the larva (689/14,426) are not expressed in the adult and, reciprocally, 39.9% (9,010/22,567) of adult genes are not expressed in the larva. Direct metacell comparisons (Fig. 2g) show that, in fact, only one larval cell type shows very strong similarity with an adult cell type: archaeocytes. Overall, this indicates that A.queenslandica larval stage deploys a unique set of cell behaviors with no counterparts in the cell types that emerge after the larva metamorphoses into adult28.
Mnemiopsis leidyi cell type diversity
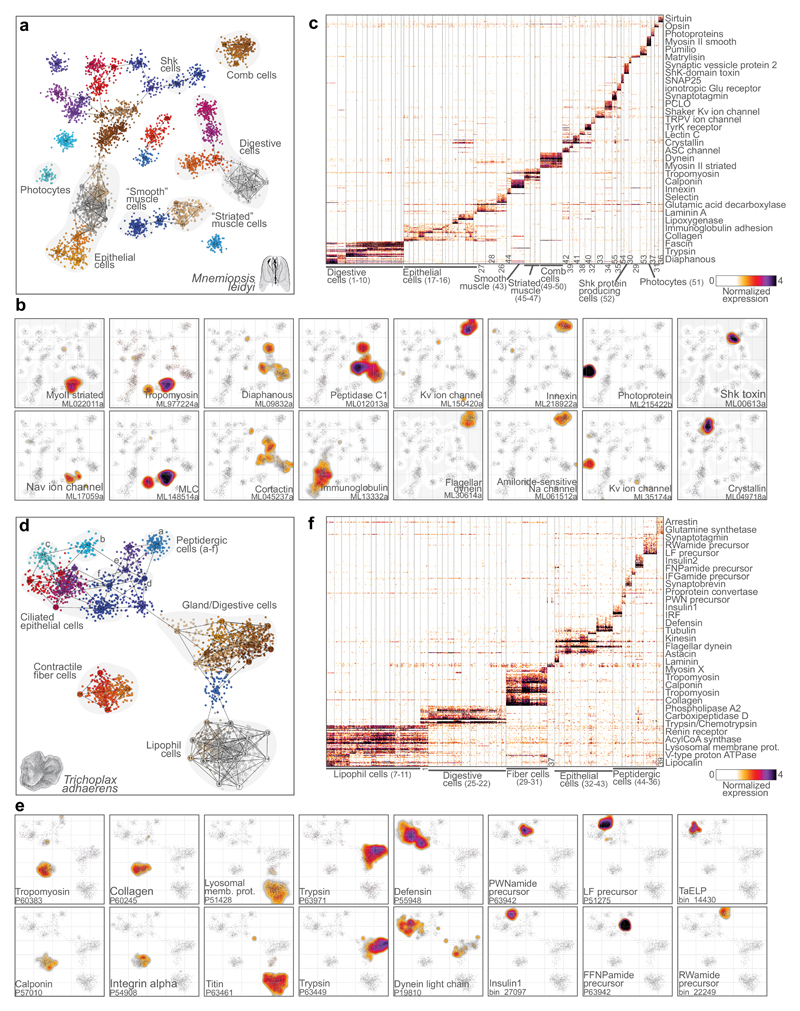
Ctenophores were traditionally considered to be sister-group to cnidarians35. However, recent phylogenomics studies clearly show they are one of the earliest-branching animal lineages, although it remains disputed whether they branch before or after sponges3–6 (Fig. 1). Ctenophores have a complex body plan and cell types like muscles and neurons. These features, together with the ctenophore phylogenetic position, open the question of whether neurons and other cell types have a single or multiple origins within Metazoa11,12,17,36. We mapped the diversity of cell types in the ctenophore M.leidyi by profiling 6,144 single cell transcriptomes. Compared to the sponge, mapping of the ctenophore M.leidyi transcriptional states uncovered a richer repertoire of cell types, some of which could be associated with putative functions and known cell types (Fig 3a-c, Supplementary Fig. 3, Table S4). For example, we could identify a group of photocyte cells (the cells responsible for ctenophore bioluminescence) expressing known photoproteins and opsins37 (Fig. 3b). Unlike most other metazoans, ctenophore locomotion is based on the coordinated ciliary beating of rows of comb cells. We identified comb cells expressing multiple ciliary markers and specific potassium voltage-gated and amiloride-sensitive sodium ion channels (Fig. 3e, Supplementary Fig. 3). Comb cells also express a specific innexin gene (Fig. 3e), supporting the existence of gap junctions electrically coupling these groups of cells, as suggested by ultrastructural observations38. Another group of cells show expression of markers associated to muscle cell types in other species39, such as tropomyosin and myosin light chain (MLC) (Fig. 3b). Interestingly, although M.leidyi lacks striated muscles, we can distinguish a group of muscle cells expressing markers associated to striated muscles in other species39 such as striated-type myosin II; while another group of muscle cells express markers of “smooth” muscles such as calponin (Fig. 3b, Supplementary Fig. 3a). We also detect cells showing expression of digestive enzymes and of genes associated to microvilli/filopodia formation40 (such as diaphanous and cortactin) (Fig. 3b); a group of cells expressing a secreted Shk-domain protein41 (Fig. 3b); and epithelial cells expressing multiple transmembrane adhesion and extracellular matrix proteins (Supplementary Fig. 3b).
Figure 3. Mnemiopsis leidyi and Trichoplax adhaerens cell type atlases.
a, 2D projection of M.leidyi metacells and single cells. b, Gene expression distribution on 2D projected M.leidyi cells for selected gene markers. c, Normalized gene expression across 4,803 M.leidyi single cells (columns), sorted by cell cluster. For each cluster, the top 25 genes sorted by fold change versus the other metacells were selected for visualization (with a FC threshold>=2). d, 2D projection of T.adhaerens metacells and single cells. e, Gene expression distribution on 2D projected T.adhaerens cells for selected gene markers. f, Normalized gene expression across 3,209 T.adhaerens single cells (columns), sorted by cell cluster. Genes selected as in c. Color-coding of cells and metacells in a and d is arbitrary.
However, most of the cell clusters we identified cannot be assigned to known functions/types and many are strongly associated to unannotated proteins (Table S3), often Ctenophora-specific (see Fig. 4e). This emphasizes our still very limited understanding of ctenophore biology9. Interestingly, we could not identify any metacell with distinct neuronal gene expression signatures such as those observed in cnidarians and bilaterians36. For example, different synaptic scaffold components are expressed across multiple cell types and no specific cell cluster shows co-expression of many voltage-gated ion channels. This lack of co-expression is similar to that observed for synaptic scaffold and other neuronal genes observed in A.queenslandica and T.adherens (see below), two organisms without neuronal cells. Instead, we find in M.leidyi highly-specific expression in multiple metacells of electrical synapse components (innexins), as well as specific expression of ASC, iGluR and Kv/Cav/Nav ion channels12,17 (Fig. 3b, Supplementary Fig. 3c-h). Overall, these findings indicate a dramatically different molecular composition of ctenophore synapses and neuronal-like cells from those of cnidarians and bilaterians, possibly suggesting convergence of these cell types12,42.
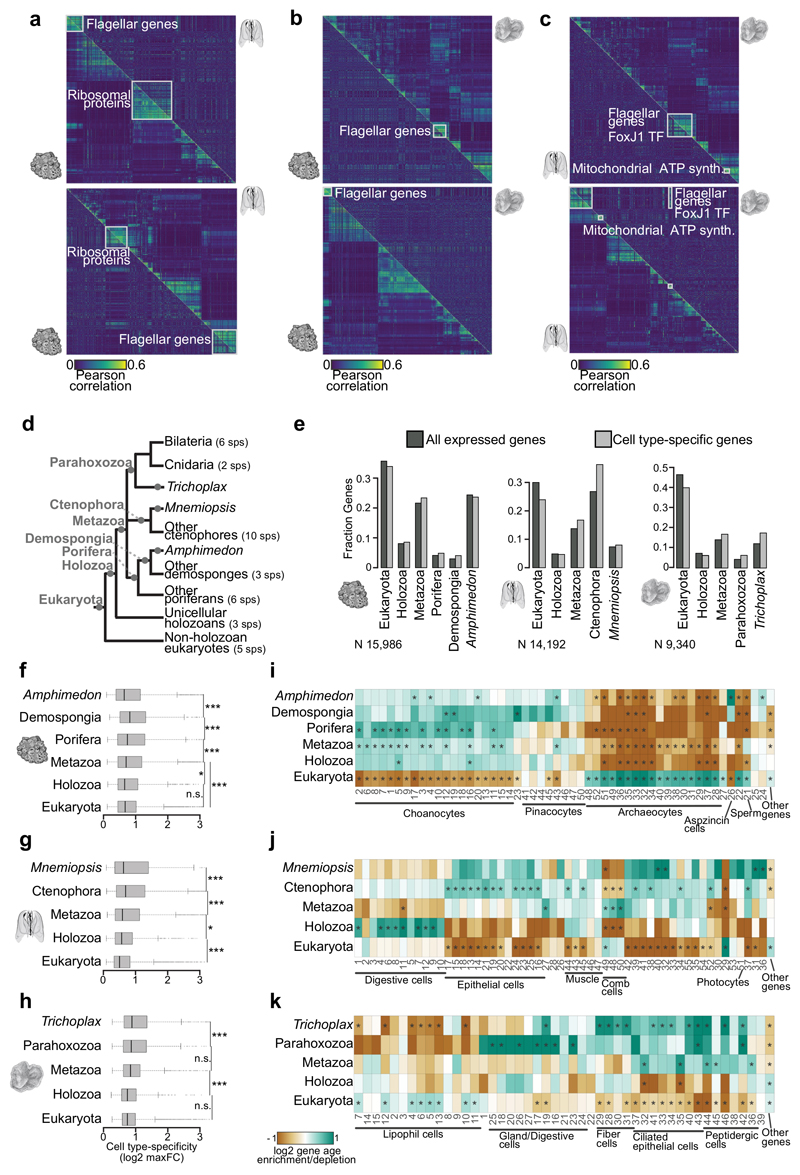
Figure 4. Phylogenetic patterns of cell type-specific genes repertoires.
a, Cross-species gene module analysis. The heatmap shows gene-gene correlation values for A.queenslandica gene modules (top panel, lower triangle) compared to the gene-gene correlation of gene orthologues in M.leidyi (sorted based on the clustering of A.queenslandica genes). Bottom panel shows the reciprocal analysis, focusing on M.leidyi gene modules and showing the equivalent correlations for A.queenslandica orthologues (lower triangle). Correlation values are computed based on expression profiles across metacells and genes are hierarchically clustered based on these correlations in the species of focus. Conserved gene modules are highlighted with white squares. b, Same as a for A.queenslandica versus T.adhaerens comparisons. c, Same as a for M.leidyi versus T.adhaerens comparisons. Notice the highly conserved flagellar toolkit gene module, found in all pairwise comparisons. Interestingly, this module is associated to FoxJ1 TF (known to be a ciliary regulator also in bilaterians58) both in M.leidyi and T.adhaerens. d, Schematic phylogenetic tree showing the lineages and number of species employed in our orthology analysis. The gene age categories derived from this analysis are shown in grey in the corresponding branches. e, Gene age distributions in each species for all detected genes (dark grey) and cell type-specific genes (light grey, genes with max fold-change >2 in at least one metacell). f, A.queenslandica genes cell type specificity (calculated as the log2 max fold-change across metacells) stratified by gene age. g, h, Same as f for M.leidyi and T.adhaerens. Notice that in all cases cell type-specificity is higher in evolutionary younger genes, with a drop in orphan/species-specific genes in the case of A.queenslandica and M.leidyi. This is in line with previous observations suggesting that gene innovations tend to be associated to tissue/cell type-specific functions47,48. *** p<<0.001, * p<0.05, n.s. non-significant; Wilcoxon rank-sum test. i, Gene age frequency enrichment/depletion in gene sets specific to each A.queenslandica metacell. The enrichment/depletion is represented as the log2 of the frequency of gene ages (among the genes overexpressed in each metacell) versus the background frequency of gene ages (taking into account only detected genes, not all predicted genes). * q-value<0.01; chi-square test (BH correction). For example, choanocytes are strongly enriched in genes of poriferan origin and, to a lesser extent, of metazoan origin. In contrast, archaeocytes and sperm cells are strongly enriched in ancient, paneukaryotic genes. j, k, Same as i for M.leidyi and T.adhaerens metacells.
Trichoplax adhaerens cell type diversity
Placozoans are the simplest (non-parasitic) multicellular animals. They have no apparent body axis or tissue-level organization, and they differentiate only 6 cell types according to ultrastructural studies14,43. These cells are organized in two ciliated epithelial layers and the flattened body is filled with extracellular matrix material and fiber cells. We dissociated and sampled the transcriptomes of 4,608 T.adhaerens cells (Fig. 3d-f, Supplementary Fig. 4, Table S5), and defined metacells and putative cell types using the same strategy than for A.queenslandica and M.leidyi. In line with the known biology and ultrastructure of T.adhaerens43, we could define groups of fiber cells, lipophil cells, digestive/gland cells, and epithelial cells, comprising together 79% of the sampled cells. Fiber cells express markers associated to cell contractility such as tropomyosin and calponin (Fig. 3e, Supplementary Fig. 4g), as well as cell adhesion and extracellular matrix proteins like integrins, collagens and fibronectins (Fig. 3e, Supplementary Fig. 4c, g). This suggests a dual role of these cells in generating the extracellular material that fill T.adhaerens body, as well as in body contraction involved, for example, in placozoan feeding behavior. Lipophil cells express multiple lysosome and lipid metabolism genes (Fig. 3e, Supplementary Fig. 4d); gland cells express different digestive enzymes like trypsins (Fig. 3e, Supplementary Fig. 4h); and epithelial cells express multiple defensins, short peptides involved in host defense (Fig. 3e, Supplementary Fig. 4e). Both gland and epithelial cells express ciliary markers (Fig. 3e, Supplementary Fig. 4f), as expected given they both are ciliated cells43.
Besides these 4 abundant cell behaviors, our analysis reveals 7 additional lower-frequency cell types, 6 of which are characterized by production of unique regulatory peptides44,45 and multiple specific transcription factors (Fig 3e, Fig. 5f). One of these regulatory peptides (TaELP, Fig. 3e) has been recently shown to regulate T.adhaerens locomotion through control of ciliary beating of the cells in the lower epithelial layer44. Therefore, we hypothesize that the 5 other peptidergic cell types we uncover in this study may be involved in the control of control additional processes, such as the release of digestive enzymes from gland cells or the contraction of fiber cells. However, although the T.adhaerens genome encode multiple genes involved in synaptic and neuronal functions18, these genes do not show co-expression in these peptidergic cell types (Supplementary Fig. 4b), indicating the absence of a synaptic scaffold or any other neuronal gene module in this placozoans. Overall, the observed states indicate that elaborated peptidic regulation occurs in this simple animal within specialized cell types that lack the characteristics of synaptic neurons44.
Phylogenetic patterns of cell type-specific genes repertoires
In order to study the evolutionary dynamics of these cell type-specific transcriptional programs, we used phylogenetic mapping to define gene ages and orthology relationships in A.queenslandica, M.leidyi and T.adhaerens (Table S6). First, we analyzed the possible cross-species conservation of cell type-specific expression correlation over orthologous gene pairs. This showed that, at the evolutionary distances separating these three species from their common ancestor (>635Ma8), co-regulation of genes is almost completely divergent (Fig. 4a-c). In fact, we only observed conserved co-regulation of specific housekeeping functions, including ribosomal proteins and flagellar apparatus.
Next we analyzed how gene age correlates with cell type transcriptional specificity (Fig. 4d-k). We defined for each gene, in each species, an inferred evolutionary origin based on the presence of orthologs in species belonging to key taxonomic groups4,12,46 (Fig. 4d, Table S6). The global age distribution among expressed genes varied substantially across species (Fig. 4e). In A.queenslandica most expressed genes are of eukaryotic origin (36%), followed by genes originated at the stem of Metazoa 23%) and A.queenslandica-specific genes (24%). In T.adhaerens paneukaryotic genes are even more dominant, representing over 50% of all expressed genes, and a similar percentage of the genes that are expressed in a cell-specific manner; while there is only a modest contribution of genes specific to T.adherens (17%) in the cell type-specific transcriptomes. In contrast with A.queenslandica and T.adherens, in the ctenophore M.leidyi most cell type-specific genes are of ctenophore origin (40%). This suggests an important contribution of ctenophore gene innovations to ctenophore cell type biology9 and also explains the difficulty of determining the identity of many of the cell clusters we identified in this species (Fig. 3a-c).
In general, genes that are expressed broadly across tissues have been shown to have older phylogenetic origins, while genes expressed in a narrower subset of tissues tend to have more recent phylogenetic origins47,48. To test if the same effect is observed in cell type transcriptomes, we defined for each gene a cell type-specificity score (based on the maximum fold-change in expression observed in any metacell) and we stratified these values according to gene age (Fig. 4f-g). In all three species, we observed that evolutionary more novel genes show significantly higher degree of cell type-specific regulation. At a higher resolution, specific cell clusters show distinct gene age distributions (Fig. 4i-k). For example, sponge choanocytes are particularly enriched in genes specific to the sponge lineage; while, in contrast, archaeocytes and sperm cells are enriched in paneukaryotic genes (Fig. 4i). In the ctenophore, digestive cells are enriched in genes of holozoan origin (ie. shared between animals and their closest unicellular relatives), while epithelial cells and multiple uncharacterized cell types are enriched in ctenophore genes (Fig. 4j). A similar pattern is observed in the placozoan T.adherens, with epithelial cells being enriched in lineage-specific genes, while lipohil cells are enriched in paneukaryotic genes and digestive cells in genes shared between placozoans, cnidarian and bilaterians (parahoxozoa) (Fig. 4k).
Cell type-specific transcription factor modules
Transcription factors (TF) are key players in the gene regulatory networks that define cell type identity49. We examined TF cell type-specific expression to test if the observed cell type transcriptional programs are linked to a rich TF repertoire. We detected expression for 168, 231, 129 predicted TFs in A.queenslandica, M.leidyi, and T.adhaerens, respectively (Fig. 5a, Supplementary Fig. 5a). The classification of predicted TFs into structural classes suggested expanded usage of homeobox and zf-C2H2 TFs in the ctenophore, but otherwise similar representation of TF classes between these species (Fig. 5b). Consistently with their likely role as key drivers of cell type regulation, we found that TFs are much more likely to be expressed in a cell type-specific fashion when compared to all other genes (Fig. 5c). Accordingly, we found different TFs being specifically expressed in all cell types in each of the species. In A.queenslandica we observed Maf, Grainyhead and 27 other TFs enriched in choanocytes; Ets and Arx homeobox are specific to pinacocytes; and Myc is expressed in archaeocytes (Fig. 5d, Supplementary Fig. 5b-c). Less frequent sponge cell types also show highly specific TF expression. For example, sperm cells show co-expression of 4 Tbx6/7 paralogs, and host defense cells express interferon regulatory factor (Fig. 5d, Supplementary Fig. 5b-c). In M.leidyi, grainyhead TF is enriched in epithelial cells and Rfx4 in the ciliated comb cells (Fig. 5e, Supplementary Fig. 6a-b). These TFs have been shown in other species to be expressed in epithelial cells and ciliated cells, respectively50,51; suggesting conserved association of these TFs with epithelial and ciliary programs. Examples of cell type-specific TF regulators in T.adhaerens include Noto homeobox in lipohil cells and FoxC in fiber cells (Fig. 5f, Supplementary Fig. 6d-e). Interestingly, while an overall similar number of TFs are expressed in a cell type-specific fashion across the three species (Fig. 2a), in the ctenophore the higher cell type complexity results in a smaller number of TFs linked with each transcriptional state, suggesting that additional epigenetic mechanisms might be involved in cell type specification for this species, for example genomic compartmentalization and combinatorial gene regulation by distal regulatory elements. In summary, elaborated combinatorial expression of TFs is observed to correlate, and possibly drive, differentiated transcriptional programs in sponges, ctenophores and placozoans.
Genomic embedding of cell type regulatory programs in early metazoans
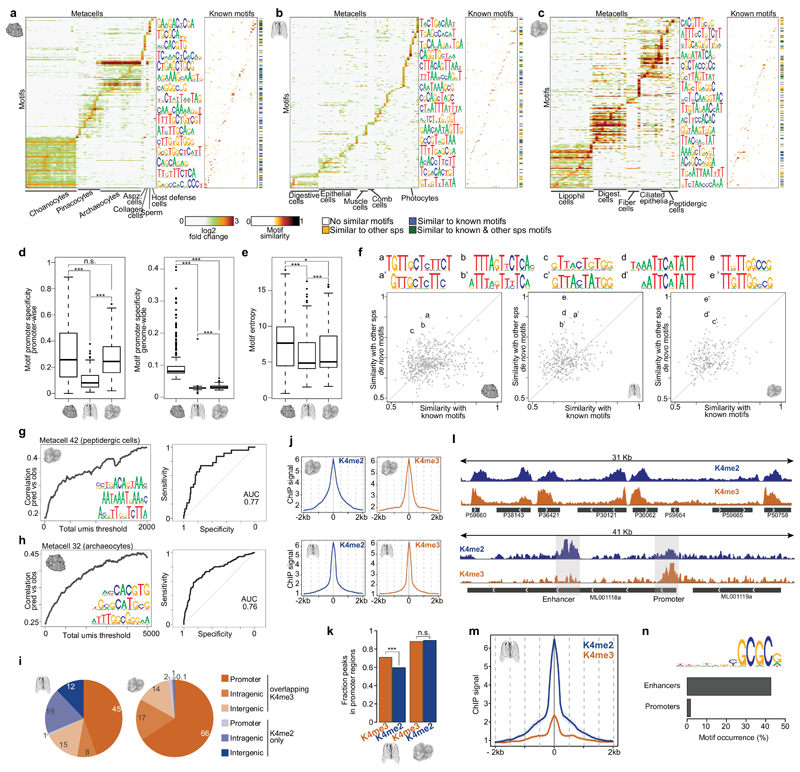
TFs regulate their target genes by binding to sequence elements located at promoters and, most prominently in bilaterians, at distal enhancers. To reconstruct the degree to which information encoded into gene promoters can direct cell type-specific transcriptional control in early metazoans, we defined sets of cell type-specific gene modules for each species (Tables S2-S5). We then searched de novo for enriched sequence motifs in predicted gene promoters (-200/+50bp from the TSS), controlling for false discovery rate and validating motif robustness by analysis of spatial motif distributions (Supplementary Fig. 7a) and shifted control sequences (Supplementary Fig. 7b). In A.queenslandica, we selected 325 motifs for downstream analysis (Table S7), computed promoter affinity to each motif, and visualized the distribution of motif enrichments for each cell type-specific gene module (Fig. 6a). This resulted in remarkably rich landscapes of promoter motif content, covering all inferred cell types with 16-96 distinct motifs. For example, we observed 93 distinct motifs enriched in choanocytes gene promoters, consistent with the exceptionally rich combination of 29 TFs associated with choanocyte-specific expression (Fig. 5d, Supplementary Fig. 5b-c). Similar analysis in M.leidyi (Fig. 6b, 6-82 motifs per cell type) and T.adhaerens (Fig. 6c, 29-98 motifs per cell type), confirmed promoter motifs are significantly enriched in these organisms as well. However, comparative analysis of the degree of motif genomic specificity (Fig. 6d) and entropy (Fig. 6e) suggested that in the ctenophore M.leidyi the strength of promoter motifs and their specificity to target genes given multiple potential genomic off-targets is significantly weaker, compared to A.queenslandica and T.adhaerens.
Figure 6. Regulatory sequence analysis in A.queenslandica, M.leidyi and T.adhaerens.
a, De novo motif enrichments in A.queenslandica promoters. Left, heatmap showing significant (FDR<0.02) motif (rows) enrichments in the promoters of metacell-specific gene sets (columns). Right, heatmap showing the similarity of each A.queenslandica promoter-enriched motif (rows) to known motifs in databases (columns). The colorbar indicates if the motif has high similarity (>0.7) with any known motifs and/or de novo motifs found in M.leidyi or T.adhaerens. b, c, Same as a for M.leidyi and T.adhaerens. d, Boxplots showing, for each species, the frequency of occurrence of metacell-specific motifs in the promoters of metacell-specific genes compared to all other gene promoters (left) and to the whole genome (right). *** p<<0.001, * p < 0.05, n.s. non-significant; Wilcoxon rank-sum test. e, Boxplot showing, for each species, the distribution of de novo motif entropies. *** p<<0.001, * p < 0.05; Wilcoxon rank-sum test. f, Scatterplots showing, for each species, the maximum similarity of each de novo motif to known motifs (x-axis) and to motifs in the other 2 species (y-axis). Highlighted cases (a-e) show examples of highly similar motifs between 2 species and not similar to any known motif in databases. g, Left, correlation between observed (obs) expression values and predicted (pred) values derived from a linear model based on promoter motif content analysis for the T.adhaerens metacell 42 (peptidergic cells). Correlation is shown as a function of the total molecule count threshold applied to the genes considered in the analysis. The three motifs with the top coefficients according to the model are shown. Right, receiver operating characteristic (ROC) curve of the linear regression model predicting gene expression in metacell 42 (peptidergic cells). h, same as g for A.queenslandica metacell 32 (archaeocytes). i, Pie charts showing the distribution of H3K4me2 peaks across different genomic feature, grouped by overlap/lack of overlap with H3K4me3 peaks. H3K4me3+K4me2 peaks in non-promoter regions are likely to represent un-annotated promoter sites. Numbers indicate the percentage of each category. j, iChIP signal metaplots centred in promoter peaks maximum coverage positions for H3K4me3 (left) and H3K4me2 (right) and in T.adhaerens (top) and M.leidyi (bottom). ChIP signal is indicated as –log2(1-coverage quantile), see Methods. k, Fraction of H3K4me2/3 peaks observed in promoter regions in M.leidyi (left) and T.adhaerens (right). *** p<<0.001; Chi-square test. l, Example T.adhaerens (top) and M.leidyi (bottom) genomic region showing normalized H3K4me2 and H3K4me3 iChIP coverage. m, M.leidyi H3K4me2/3 iChIP signal metaplots centred at enhancer elements maximum H3K4me2 positions. n, De novo motif enriched in M.leidyi enhancers. Barplot shows the frequency of occurrence of this motif in M.leidyi enhancers and promoters.
Our de novo discovery approach is a priori not restricted to identification of known TF binding motifs characterized in model species. Nevertheless, we found that 33% of A.queenslandica, 25% of M.leidyi and 32% of T.adhaerens motifs matched (similarity >0.7) known models retrieved from databases covering TF motifs for multiple eukaryotic species (Fig. 6a-c, Supplementary Fig. 7f). This indicates that at least some of the sequence elements defining the TF-genome interface are deeply evolutionary conserved. Remarkably, out of the 570 novel motifs that could not be matched in databases, we detected 53 conserved between at least two species (Fig 6f, Supplementary Fig. 7g). Discovering novel motifs independently in highly diverged species serves as further validation of the robustness of the promoter signals we characterize and indicate that comprehensive characterization of the repertoire of possible TF-DNA interfaces in metazoan genomes will require further analysis of phylogenetically diverse species.
Analysis of promoter information content by predictive expression models
In multicellular animals stable differentiated transcriptional programs are defined by multiple cis-regulatory modules, long range control and powerful epigenetic mechanisms52. In contrast, in most unicellular eukaryotes gene regulation involves exclusively regulatory elements that are proximal to the gene promoter53. Hence, we were surprised by the high degree of proximal promoter information content in A.queenslandica and T.adhaerens. To further quantify this information content in cell type-specific promoters, we implemented a simple model aiming to predict cell type-specific expression from promoter sequences alone (see Methods). We tested the model by training on subsets of the genes and then predicting cell type-specific gene expression from hidden promoter sequences. We found that this simple approach generated substantial predictive value in multiple A.queenslandica and T.adhaerens metacells (Fig. 6g-h, Supplementary Fig. 7c-e), despite the clear limitations of predicting combinatorial regulation using linear models. Accuracy improved as the total number of RNA molecules captured for a gene was increasing (Fig. 6g-h, Supplementary Fig. 7c-e), indicating some of the inaccuracy of our predictions stems from experimental noise in the estimation of differential expression. For example, using promoter sequences alone, we could predict 50% of the A.queenslandica metacell 32 gene expression with 90% specificity (AUC = 0.76) and 50% of the T.adhaerens metacell 42 gene expression with 84% specificity (AUC=0.77). Interestingly, predictions based on promoter sequence were less powerful in the ctenophore (Supplementary Fig. 7d), suggesting important contribution of additional, perhaps distal, regulatory elements in this group.
Characterizing distal epigenetically marked loci in Mnemiopsis leidyi
To test the potential contribution of long-range regulatory elements in M.leidyi and, as a control, in T.adhaerens, we used iChIP54 in these two species. We profiled chromatin extracted from whole organisms with antibodies against histone modifications associated with promoter (H3K4me2/3) and enhancer (H3K4me2-only) activities. We found that whole organism iChIP was sufficiently sensitive to detect H3K4me2/3 enrichment in 45% of M.leidyi and 66% of T.adhaerens promoters (Fig. 6i), showing quantitatively stronger enrichment for promoters that were expressed in a larger fraction of the cells (Supplementary Fig. 8a-b). Spatial analysis showed H3K4me3 and H3K4me2 are localized around annotated promoters in distance scale of less than 500bp in both species (Fig. 6j). Interestingly, we found that while in T.adhaerens the fraction of H3K4me2 and H3K4me3 peaks mapping in promoter regions are the same (Fig. 6k), a significant fraction of H3K4me2 in M.leidyi do not co-localize with H3K4me3 in promoters, suggesting the existence of non-promoter distal regulatory elements54. Examples of epigenomic profiles (Fig. 6l, Supplementary Fig. 8c-d) and spatial mapping around distal H3K4me2 in the ctenophore (Fig. 6m) both support the existence of a distinct class of distal epigenetically marked loci in this species. Furthermore, sequence analysis revealed that these loci are 20-fold enriched for a specific GCGC-rich motif compared to promoters (5-fold compared to the genomic background) (Fig. 6n, Supplementary Fig. 8e-f). The strong chromatin signature we observe in whole-organism ChIP-seq for these class of distal elements and the strong sequence specificity observed in it suggest this class represent some constitutively active genomic-structural elements. Such element may be hypothesized to perform functions that are similar to the role of CTCF in vertebrates55 or of Beaf-32 in Drosophila melanogaster56,57. In summary, we discovered the existence of distal elements M.leidyi with strong sequence specificity and a potential role as enhancers and/or chromosomal organizers. Similar analysis could not detect any evidence for distal regulatory elements in T.adhaerens.
Discussion
Using whole organism single-cell RNAseq and a combination of sequence and chromatin analysis, we mapped differentiated transcriptional states and linked them with putative cell types in three representatives of the earliest-branching animal lineages. The unbiased approach we employed provides the first systematic insight into early animal cell type regulatory programs, revealing distinct cell type repertoires in sponge adult and larva, a surprisingly high diversity of cell types in M.leidyi, and the existence of multiple specialized peptidergic cell types in T.adhaerens. Combination of this cell type transcriptional atlases with chromatin and sequence analyses indicates the existence of some key differences between the sponge, placozoan and ctenophore cell type-specific transcriptional control schemes. On the one hand, A.queenslandica and T.adhaerens have fewer cell types and show remarkably specific promoter sequence motifs. Moreover, T.adhaerens shows no evidence of regulation by distal enhancer elements. On the other hand, M.leidyi has higher cell type diversity, expresses fewer specific TFs per cell type, and shows lower information content in gene promoters. Moreoever, M.leidyi shows strong evidence for distal regulatory elements. We suggest that the ctenophore mechanistic solution for defining and stabilizing cell types programs might be more similar to the bilaterian solution, employing multiple layers of control to supplement the transcription factor combinatorics. We hypothesize that this elaborate regulation might be necessary to specify large repertoires of cell types embedded in a complex bodyplan such as that of ctenophores. In contrast, placozoans demonstrate the feasibility of defining and regulating multiple cell types without such strong layered architecture, but simply using a combination of TFs and proximal promoter regulatory elements, similarly to what is observed in unicellular eukaryotes and unlike the animal species studied to date. We expect the methodology we introduce here will facilitate multiple studies for mapping cell type regulation in diverse species in the coming years, resulting in an increasingly dense phylogenetic coverage of cellular behaviors across the animal tree of life. The integrative analysis of this data will further allow a comprehensive and principled analysis of the evolutionary mechanisms leading to animal multicellularity and the genomic determinants of multifaceted transcriptional control schemes.
Methods
Animal sources, specimen dissociation and cell sorting
Amphimedon queenslandica adults and larvae were collected from Heron Island Reef, Great Barrier Reef, Queensland, Australia. Adult specimens were dissociated by placing them in a syringe and squeezing them through a 60μm nylon mesh (fused to the end of the syringe) into calcium/magnesium-free seawater (CMFSW). Larvae were dissociated by gentle pipetting with gelatin-coated tips.
Mnemiopsis leidyi adults originated from L. Friis-Møller, Kristineberg, Sweden. They were maintained in the lab in filtered seawater, with small adult specimens (~20 mm) used for dissociation. Specimens were starved for 2-3 days, with daily changes of seawater. They were relaxed briefly in 7% magnesium chloride, then rinsed twice in CMFSW. For dissociation, they were incubated in 0.25% chymotrypsin (MP Biomedicals) in CMFSW for 20 min at room temperature with constant rocking and gentle pipetting. Cells were collected by centrifugation for 10 min at 1,000g at 16C.
Trichoplax adhaerens (Grell strain60) were cultured in the laboratory at room temperature, using artificial seawater (ASW) and feeding them with the cryptophyte algae Pyrenomonas helgolandii (strain SAG 28.87). Algae were obtained from University of Gottingen algae culture collection (SAG), and cultured at room temperature in 250ml flasks using PROV50 medium (#MKPROV50L, NMCA) and a long wavelength fluorescent lamp. For dissociation, 30-40 animals were first transferred to a small plastic dish and, after they attached, cleaned 3x with ASW. Then, ASW was replaced by CMFSW+10mM EDTA and animals were dissociated by gentle pipetting with gelatin-coated tips.
In all cases, cells were distributed into 384-wells capture plates (all coming from the same production batch) containing 2ul of lysis solution using a using a FACSARIA III cell sorter. Lysis solution contain 0.2% Triton and RNAse inhibitors plus barcoded poly(T) reverse-transcription (RT) primers for single cell RNAseq. Non-cellular particles were discriminated by selecting only DRAQ5-positive cells (25uM DRAQ5 staining, Thermo #62251) and cell doublet/multiplet exclusion was performed using FSC-W versus FSC-H. Fresh cell dissociates were prepared every 2h and sorted plates were immediately spun down, to ensure cell immersion into the lysis solution, and frozen at -80ºC until further processing.
Massively Parallel Single-Cell RNAseq (MARS-seq)
Single cell libraries were prepared as previously described22. For each species, all single cell libraries were prepared in parallel: 8,832 libraries for A.queenslandica (13 plates for adult sponges and 10 for larvae), 6,144 for M.leidyi (16 plates) and 4,224 for T.adhaerens (12 plates). That is, we employed exactly the same conditions (incubation times, temperatures, etc.) and reagents, in order to minimize technical factors. First, using a Bravo automated liquid handling platform (Agilent), mRNA was converted into cDNA with an oligo containing both the unique molecule identifiers (UMIs) and cell barcodes. Unused oligonucleotides were removed by Exonuclease I treatment. cDNAs were pooled (each pool representing half of the original 384-wells MARS-seq plate) and linearly amplified using T7 in vitro transcription and the resulting RNA was fragmented and ligated to an oligo containing the pool barcode and Illumina sequences, using T4 ssDNA:RNA ligase. Finally, RNA was reverse transcribed into DNA and PCR amplified. Resulting libraries were tested for amplification using qPCR and the size distribution and concentration were calculated using Tapestation (Agilent) and Qubit (Invitrogen). For each species, all scRNAseq libraries were pooled at equimolar concentration and sequenced to saturation (>=4 reads/UMI) using Illumina NextSeq 500 sequencer and using a mid-output 75 cycles v2 kit (Illumina). For adult A.queenslandica, we obtained a total of 430M reads, with an average depth of 53,000 reads per cell and 6 reads/UMI on average (Table S1). For A.queenslandica larvae, we obtained a total of 67M reads, with an average depth of 11,000 reads per cell and 5 reads/UMI on average. For M.leidyi, we obtained a total of 506M reads, with an average depth of 36,000 reads per cell and 5 reads/UMI on average. In the case of T.adhaerens, we obtained a total of 85M reads, with an average depth of 14,000 reads per cell and 7 reads/UMI on average.
MARS-seq reads processing and filtering
Reads were mapped into A.queenslandica, T.adhaerens and M.leidyi genomes using bowtie2 (with parameters: -D 200 -R 3 -N 1 -L 20 -i S,1,0.50) and associated with gene intervals. For each species, we extended gene intervals up to 2kb downstream or until the next gene in the same strand is found. This accounts for the poor 3'UTR annotation of these species, which causes many of the MARS-seq (a 3' biased RNAseq method) reads to map outside genes. Additionally, in order to account for putative unannotated genes, we defined 500bp bins (not covered by our gene intervals) genome-wide. We retained those with >=10 uniquely mapping reads and used them in the cell clustering process (see below).
Mapped reads were further processed and filtered as previously described22. UMI filtering include two components, one eliminating spurious UMIs resulting from synthesis and sequencing errors, and the other eliminating artifacts involving unlikely IVT product distributions that are likely a consequence of second strand synthesis or IVT errors. The minim FDR q-value required for filtering was 0.2.
Metacell and clustering analysis
We used the MetaCell package (Appendix S1) to select gene features, construct gene modules and create projected visualization of the data, using parameters as described below. The complete analysis code is available at https://dl.dropboxusercontent.com/s/n36cusnenvi306a/Code_and_data.tar.gz?dl=0. We applied preliminary cell filtering based on total UMI counts using a permissive threshold of 100 UMIs (50 UMIs in the case of A.queenslandica larva, to account for the very different molecule count distributions in this sample). For gene selection we used a normalized depth scaling correlation threshold of -0.1 (-0.05 in A.queenslandica larva and T.adhaerens), and total UMI count of more than 100 molecules (empirical median marker UMI count was 2,723 for the sponge, 1,013 for the ctenophore, 1,075 for the placozoan). For metacell construction we used K=150, minimum module size of 30, and automatic filtering of background noise using an initial epsilon value of 0.03. Bootstrapping was performed using 1,000 iterations of resampling 75% of the cells, leading to estimation of co-clustering between all pairs of single cells and identification of robust clusters based on single or grouped metacells. For 2D projections, in A.queenslandica adult dataset we used a K-nn constant of 50, and restricted the module graph degree by at most 10 (A.queenslandica larva, K=30/max degree=3; M.leidyi K=30/max degree=7; T.adhaerens K=30/max degree=8).
We performed manual validation and adjustment of the automatic module covers in Fig. 1 and Supplementary Fig. 4 as follows. We filtered metacells that were not enriched by at least three genes at over 3 fold over the median of the entire populations. Additionally, module-specific transcriptional enrichment was tested for each metacell by identifying a set of module-specific genes (top 50 genes with FC>=2) and computing the top 1% of their total expression across all non-module cells (excluding also cells in the two most similar modules). Given this top percentile as a threshold, the fraction of cells in the module that express the module’s genes over the threshold was computed, and additional module filtering was applied if this value was lower than 30%. We also filtered out metacells with less than 10,000 total molecules. We note that cells that were filtered during this combined scheme may be part of additional undetected states, or may represent weaker signal that is in fact part of other, more robust modules, but that for our goals in the analysis here, robustness of the reported transcriptional states and the subsequent genomic analyses is key. Overall this resulted in filtering 862 cells in the sponge, 785 cells in the ctenophore and 188 cells in the placozoan. Finally, we merged metacells with >20% shared cells co-clustering in our 1,000 bootstrap replicates were merged, resulting in the metacells presented in Fig. 2 and Fig. 3, and supported by bootstrap analysis in Supplementary Fig. 1.
Indexing-first Chromatin Immunoprecipitation (iChIP)
For iChIP experiments, M.leidyi and T.adhaerens cells (dissociated as described above) were crosslinked in 1% formaldehyde for 10 min at room temperature (RT). Crosslinking was quenched with 0.125M glycine for 5min RT. Cross-linked cells were pelleted and stored at -80C. Chromatin was sonicated in a Bioruptor sonicator (Diagenode), distributing 1M cells/100ul tube and using 45 sonication cycles (30” ON/30” OFF, High mode). Then, chromatin was immobilized onto anti-H3 antibody (Abcam, #ab1791) coated Protein G Beads (Invitrogen). After 3 washes with 10mM Tris pH8 + protease inhibitors (PI), immobilized chromatin was indexed with Illumina Y-shapped adaptors as described in54. After barcoding, indexed chromatin was pooled and released from Ab-ProtG bead immuno-complexes by incubating 30min at 37C in a buffer containing 50mM EDTA, 2% SDS, 2% Deoxycholic acid and 1M NaCl. After the incubation, the chromatin was separated from the magnetic beads using a magnet and the released indexed chromatin was transferred to another tube and diluted 1 to 20 in a buffer of 10mM TrisCl, 10mM NaCl, 1mM EDTA. A small fraction of this dilution (60ul) was separated to be sequenced as input. The remaining diluted indexed chromatin (approx. 10ml) was concentrated to 200ul using a 50Kda centricon (Ambion) and the volume was brought to 400ul with RIPA buffer + PI. The 400ul pool was divided in 2 to perform two ChIP assays, one for H3K4me2 and another for H3K4me3. The specific ChIP reaction was carried out at this stage by incubating the 200ul extract of indexed chromatin pool with 4ul of anti-H3K4me2 antibody (Abcam, #ab3236) or 2.5ul of anti-H3K4me3 antibody (Millipore, #07-473) at 4C with rotation. After 10 h of incubation, 40ul of pre-washed ProtG beads were added and incubated for 1h to capture the Ab-chromatin complexes. Immunocomplexes were then washed 5X with RIPA (150mM NaCl, 0.1% SDS, 0.1% Deoxycholate, 1% Tx-100, 1mM EDTA), 2X with RIPA-500 (500mM NaCl, 0.1% SDS, 0.1% Deoxycholate, 1% Tx-100, 1mM EDTA), 2X with LiCl buffer (250mM LiCl, 0.5% NP-40, 0.5% Deoxycholate, 1mM EDTA), 2X with TE, and resuspended in 50ul of Chromatin Elution Buffer (0.4% SDS, 250mM NaCl, 5mM EDTA, 10mM TrisCl ph 8) plus 2.5ul of Proteinase K (NEB) and incubated for 2h at 37C and 6h at 65C. ChIPed DNA was purified with AMPure beads with a ratio of 2.5X and eluted in 23ul of EB (10mM Tris pH8). 12 cycles of PCR were performed to amplify the ChIPed barcoded DNA using 25ul of 2X HiFi Kappa Master Mix and 2ul of primer master mix.
iChIP libraries were sequenced using Illumina NextSeq 500 sequencer. For M.leidyi, the total number of reads was: 21M (H3K4me2), 12M (H3K4me3) and 10M (input). For T.adhaerens, the total number of reads was: 24M (H3K4me2), 14M (H3K4me3), and 11M (input).
iChIP analysis and enhancer definition
iChIP reads trimmed to 37nt and then mapped into the corresponding reference genome using Bowtie v1.1.161 with -v 3 -m 1 parameters. Duplicates reads were removed using SAMtools v1.162. Mapped reads were extended to 200bp (iChIP libraries fragment size) and 1bp-resolution coverage statistics over each of the genomes were computed.
To control for ChIP-seq coverage and variable ChIP-seq specificity, we transformed raw coverage values to quantile values. H3K4me3 and H3K4me2 peaks were defined as regions with coverage quantiles over 0.97 (in M.leidyi) or 0.94 (in T.adhaerens), merging peaks located at <200bp. To account for mappability/assembly problems (e.g. repetitive regions), we defined “peaks” using input data and excluded those regions from our H3K4me3/me2 peaks. In downstream analysis, iChIP coverage is indicated as –log2(1-coverage quantile), in a way that, for example, a normalized value of 9 indicates coverage is in the top 1-2-9 quantile (in the top 1/512th of the distribution).
H3K4me3 is associated to promoter elements while H3K4me2 is associated to both promoters and enhancers54. We used this property to search for distal enhancer elements in M.leidyi and T.adhaerens, by asking for H3K4me2 peaks that are >=2Kb from any H3K4me3 or any TSS (of an expressed gene, >=5 total UMIs detected).
Sequence motif analysis
We extracted promoter sequences using -200/+50 bp from annotated TSSs and associated sequences with metacells whenever their gene was at least two-fold over-expressed in the module compared to the background. We then performed de novo motif enrichment analysis for the regulatory sequences associated to each gene list, using Homer findMotifsGenome.pl (with default parameters, searching for 25 motifs and with a constant fragment size of 250bp)63. For each species, we grouped all the resulting de novo motifs and we used Homer compareMotif.pl to filter motifs (min p-value <1e-10, min number of hits in target sequences >=10) and then merge redundant motifs (>0.8 similarity threshold). Additionally, in the case of M.leidyi, we searched for enriched motifs in all enhancers (1,157) vs all genome, using a homer fragment size of 600bp.
For comparison of de-novo motifs with the database, we used data from Jolma et al.64,65, HocoMoco database63,66, JASPAR, Drosophila DPMMPMM, plant AthaMap, and Saccharomyces motif collections from Harbinson et al. and MacIsaac et al. . We computed similarities between motifs (Fig. 6, Supplementary Fig. 7) using the motifSimilarity function of PWMenrich R library, which computes the normalized sum of correlations between motif position frequency matrices.
As a result of the de novo motif finding, filtering and merging, we obtained a single set of motifs per each species. We then analysed the overrepresentation of specific motifs in promoters associated to metacell-specific gene modules. For a short sequence element s[1..k] = s1,..,sk, and a PWM wi[c], the standard local probability model is defined by multiplication: log(P(s)) = \sumilog(wi[si]) and the binding energy for a larger sequence element can be approximated 67 by E(s[1..n]) = log(\sum(j = 1:(n-k))P(s[j:(j+k)]). For each PWM, the 0.98 quantiles of genome-wide binding energies in windows of 250bp (same size as promoters) were determined. These quantiles values were then used as thresholds to determine motif occurrence for each PWM at each element. The enrichment level of each PWM/metacell pair was computed as the fold change between the frequency of occurrence of a motif in the metacell promoters and the frequency in the background gene set (all other genes detected in this study). Enrichments were assessed statistically using a hypergeometric test. We account for multiple testing by performing 100 random permutations of the promoter-motif energy matrix, computing p-values for each permutation and using the resulted distribution to derived FDR values on the empirical enrichments. An FDR threshold of 0.02 was used for the motif enrichment visualization. Additionally, only motifs with a fold change enrichment over 1.5 in at least one metacell, and a minimum foreground count of 5 (i.e. at least five genes in the metacell gene set with the motif in their promoters) and a background count of 100 were considered.
Finally, we performed cross-validation analysis by dividing expressed genes into 5 blocks and, for each of them, run the whole de novo motif discovery pipeline with the other 80% of the genes (training set). Using the glmnet R package, we built a Lasso regularized linear model based on the promoter motif energies and gene expression values of the training set (80%). We then employed this model to predict the expression values of the gene test set (20%) based on the motif energies in their promoters. We did this for each of the 5 blocks, resulting in predicted expression values for all expressed genes in our dataset. ROC curves and AUC values were computed using the pROC R package.
Gene functional annotation
We used blastp (with parameters –evalue 1e-5 and -max_target_seqs 1) to find the most similar, if any, human, fruit fly and yeast homologs (retrieved from Uniprot) for each protein of the predicted A.queenslandica, M.leidyi and T.adhaerens predicted proteomes. Additionally, we predicted for each protein the Pfam domain composition using Pfamscan68 with default curated gathering threshold. TFs were identified using univocal Pfam domains for each structural TF family69. In the case of multi-TF families (Homeobox, Fox, bHLH, bZIP, DM, Smad, Myb, NR, RFX, RHD, SRF, Ets, T-box and Sox), we used phylogenetic analyses for each family to classify them into specific subfamilies (together with the complete TF sets of additional 10 animal species, including Homo sapiens and Drosophila melanogaster for reference annotation). Briefly, sequences were aligned using MAFFT70, the resulting analysis were manually edited, ProtTest71 was used to define the best-fit aminoacidic substitution model in each case, and then phylogenies were computed using RAxML72 and Phylobayes73, for maximum likelihood and Bayesian inference, respectively. We used a similar strategy to build a phylogeny of A.queenslandica aspzincins (Supplementary Fig. 2e), extending our search for aspzincins to other eukaryotic and bacterial species. To this end, we used the presence of Aspzincin_M35 domain (PF14521, Pfam) to identify aspzincins in different species.
Phylogenetic distribution and gene age estimation
We used the complete predicted proteomes of 39 species (Table S6) at key phylogenetic positions in order to compute orthogroups, including an extensive set of 11 ctenophore species (Beroe abyssicola, Bolynopsis infundibulum, Coeloplana astericola, Coeloplana meteoris, Dryodora glandiformis, Euplokamis dunlapae, Mertensiidae sp, Vallicula multiformis, Lampea pancerina, Pleurobrachia bachei, Mnemiopsis leyidi)4,12, 10 poriferan species (Clathrina coriacea, Grantia compressa, Leuconia nivea, Sycon ciliatum, Plakina jani, Oscarella carmela, Pleraplysilla spinifera, Amphimedon queenslandica, Eunapius carteri, Ephydatia muelleri)4,46, the placozoan Trichoplax adhaerens, 10 cnidarian+bilaterian species (Homo sapiens, Branchiostoma floridae, Drosophila melanogaster, Tribolium castaneum, Capitella teleta, Lottia gigantean, Acropora digitifera, Nematostella vectensis), and 8 non-metazoan eukaryotes (Salpingoca rosetta, Capsaspora owczarzaki, Creolimax fragrantissima, Saccharomyces cerevisiae, Spizellomyces punctatus, Dictyostelium discoideum, Arabidopsis thaliana, Naegleria gruberi). We computed reciprocal blast results between all complete proteomes, with fixed database size and e-value threshold of 1e-04. Based on these reciprocal blast results, orthogroups were computed using orthoMCL algorithm74 with an inflation value (I parameter) of 1.3. We parsed these orthogroups using a parsimony criterion in order to generate an age estimation for each A.queenslandica, M.leidyi and T.adhaerens gene.
Supplementary Material
Acknowledgments
We thank all the members of the Tanay lab for comments and discussion, Xavier Grau-Bové for genome statistics in different species, Anlaug Furu for help with M.leidyi, and Hans-Jürgen Osigus and Prof. Bernd Schierwater for providing T.adharens starting culture. Research in A.H. group was supported Sars Core Budget (K.P., A.H.) and an NSF IRFP Postdoctoral Fellowship (1158629) to K.P. Research by B.M.D. is supported by the Australian Research Council. A.S.-P. was supported by an EMBO Long-Term Fellowship (ALTF 841-2014). Research in A.T. group was supported by the European Research Council. A.T. is a Kimmel investigator.
Footnotes
Data Availability
All data was deposited in GEO with the accession number GSE111068. The MetaCell package, UMI tables and annotation files are available on our group website: http://compgenomics.weizmann.ac.il/tanay/?page_id=99
Competing interests
The authors declare no competing financial interests.
Author Contributions
A.S.-P. and A.T. conceived the project. K.P., A.H., B.M.D. and F.D. provided animal specimens and chromatin material. A.S.-P. performed MARS-seq experiments. A.S.-P. and D.L. performed iChIP experiments. A.S.-P. and A.T. analysed the data and wrote the manuscript. All authors discussed and commented on the data.
References
- 1.Arendt D, et al. The origin and evolution of cell types. Nat Rev Genet. 2016;17:744–757. doi: 10.1038/nrg.2016.127. [DOI] [PubMed] [Google Scholar]
- 2.Sebé-Pedrós A, Degnan BM, Ruiz-Trillo I. The origin of Metazoa: a unicellular perspective. Nat Rev Genet. 2017;18:498–512. doi: 10.1038/nrg.2017.21. [DOI] [PubMed] [Google Scholar]
- 3.Whelan NV, et al. Ctenophore relationships and their placement as the sister group to all other animals. Nat Ecol Evol. 2017;1:1737–1746. doi: 10.1038/s41559-017-0331-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Simion P, et al. A Large and Consistent Phylogenomic Dataset Supports Sponges as the Sister Group to All Other Animals. Curr Biol. 2017;27:958–967. doi: 10.1016/j.cub.2017.02.031. [DOI] [PubMed] [Google Scholar]
- 5.Hejnol A, et al. Assessing the root of bilaterian animals with scalable phylogenomic methods. Proc R Soc London Ser B, Biol Sci. 2009;276:4261–70. doi: 10.1098/rspb.2009.0896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dunn CW, et al. Broad phylogenomic sampling improves resolution of the animal tree of life. Nature. 2008;452:745–749. doi: 10.1038/nature06614. [DOI] [PubMed] [Google Scholar]
- 7.Valentine JW. In: Keywords and Concepts in Evolutionary Devlopmental Biology. Hall B, Olson W, editors. Harvard University Press; 2003. pp. 35–53. [Google Scholar]
- 8.Cunningham JA, Liu AG, Bengtson S, Donoghue PCJ. The origin of animals: Can molecular clocks and the fossil record be reconciled? BioEssays. 2017;39:e201600120. doi: 10.1002/bies.201600120. [DOI] [PubMed] [Google Scholar]
- 9.Dunn CW, Leys SP, Haddock SHD. The hidden biology of sponges and ctenophores. Trends Ecol Evol. 2015;30:282–291. doi: 10.1016/j.tree.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 10.Jager M, Manuel M. Ctenophores: an evolutionary-developmental perspective. Curr Opin Genet Dev. 2016;39:85–92. doi: 10.1016/j.gde.2016.05.020. [DOI] [PubMed] [Google Scholar]
- 11.Jákely G, Paps J, Nielsen C. The phylogenetic position of ctenophores and the origin(s) of nervous systems. Evodevo. 2015;6:1. doi: 10.1186/2041-9139-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moroz LL, et al. The ctenophore genome and the evolutionary origins of neural systems. Nature. 2014;510:109–114. doi: 10.1038/nature13400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Simpson TL. The Cell Biology of Sponges. Springer; New York: 1984. [Google Scholar]
- 14.Schierwater B, DeSalle R. Placozoa. Curr Biol. 2018;28:R97–R98. doi: 10.1016/j.cub.2017.11.042. [DOI] [PubMed] [Google Scholar]
- 15.Srivastava M, et al. The Amphimedon queenslandica genome and the evolution of animal complexity. Nature. 2010;466:720–726. doi: 10.1038/nature09201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fernandez-Valverde SL, Calcino AD, Degnan BM. Deep developmental transcriptome sequencing uncovers numerous new genes and enhances gene annotation in the sponge Amphimedon queenslandica. BMC Genomics. 2015;16:387. doi: 10.1186/s12864-015-1588-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ryan JF, et al. The Genome of the Ctenophore Mnemiopsis leidyi and Its Implications for Cell Type Evolution. Science. 2013;342 doi: 10.1126/science.1242592. 1242592–1242592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Srivastava M, et al. The Trichoplax genome and the nature of placozoans. Nature. 2008;454:955–960. doi: 10.1038/nature07191. [DOI] [PubMed] [Google Scholar]
- 19.Putnam NH, et al. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science. 2007;317:86–94. doi: 10.1126/science.1139158. [DOI] [PubMed] [Google Scholar]
- 20.Cao J, et al. Comprehensive single-cell transcriptional profiling of a multicellular organism. Science. 2017;357:661–667. doi: 10.1126/science.aam8940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sebe-Pedros A, et al. Cnidarian cell type diversity revealed by whole-organism single-cell RNA-seq analysis. bioRxiv. 2017 doi: 10.1101/201103. [DOI] [PubMed] [Google Scholar]
- 22.Jaitin DA, et al. Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science. 2014;343:776–9. doi: 10.1126/science.1247651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gonobobleva E, Maldonado M. Choanocyte ultrastructure in Halisarca dujardini (Demospongiae, Halisarcida) J Morphol. 2009;270:615–27. doi: 10.1002/jmor.10709. [DOI] [PubMed] [Google Scholar]
- 24.Funayama N, Nakatsukasa M, Hayashi T, Agata K. Isolation of the choanocyte in the fresh water sponge, Ephydatia fluviatilis and its lineage marker, Ef annexin. Dev Growth Differ. 2005;47:243–253. doi: 10.1111/j.1440-169X.2005.00800.x. [DOI] [PubMed] [Google Scholar]
- 25.Nickel M, Scheer C, Hammel JU, Herzen J, Beckmann F. The contractile sponge epithelium sensu lato - body contraction of the demosponge Tethya wilhelma is mediated by the pinacoderm. J Exp Biol. 2011;214:1692–1698. doi: 10.1242/jeb.049148. [DOI] [PubMed] [Google Scholar]
- 26.Nichols SA, Roberts BW, Richter DJ, Fairclough SR, King N. Origin of metazoan cadherin diversity and the antiquity of the classical cadherin/β-catenin complex. Proc Natl Acad Sci U S A. 2012;109:13046–51. doi: 10.1073/pnas.1120685109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Adamska M, et al. The evolutionary origin of hedgehog proteins. Curr Biol. 2007;17:836–837. doi: 10.1016/j.cub.2007.08.010. [DOI] [PubMed] [Google Scholar]
- 28.Nakanishi N, Sogabe S, Degnan BM. Evolutionary origin of gastrulation: insights from sponge development. BMC Biol. 2014;12:26. doi: 10.1186/1741-7007-12-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Müller WEG. The stem cell concept in sponges (Porifera): Metazoan traits. Semin Cell Dev Biol. 2006;17:481–491. doi: 10.1016/j.semcdb.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 30.Alié A, et al. The ancestral gene repertoire of animal stem cells. Proc Natl Acad Sci. 2015;112:E7093–E7100. doi: 10.1073/pnas.1514789112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rieger RM. The Biphasic Life Cycle—A Central Theme of Metazoan Evolution. Am Zool. 1994;34:484–491. [Google Scholar]
- 32.Degnan SM, Degnan BM. The origin of the pelagobenthic metazoan life cycle: what’s sex got to do with it? Integr Comp Biol. 2006;46:683–690. doi: 10.1093/icb/icl028. [DOI] [PubMed] [Google Scholar]
- 33.Nakanishi N, Stoupin D, Degnan SM, Degnan BM. Sensory Flask Cells in Sponge Larvae Regulate Metamorphosis via Calcium Signaling. Integr Comp Biol. 2015;55:1018–1027. doi: 10.1093/icb/icv014. [DOI] [PubMed] [Google Scholar]
- 34.Adamska M, et al. Wnt and TGF-β Expression in the Sponge Amphimedon queenslandica and the Origin of Metazoan Embryonic Patterning. PLoS One. 2007;2:e1031. doi: 10.1371/journal.pone.0001031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Philippe H, et al. Phylogenomics revives traditional views on deep animal relationships. Curr Biol. 2009;19:706–12. doi: 10.1016/j.cub.2009.02.052. [DOI] [PubMed] [Google Scholar]
- 36.Liebeskind BJ, Hofmann HA, Hillis DM, Zakon HH. Evolution of Animal Neural Systems. Annu Rev Ecol Evol Syst. 2017;48:377–398. [Google Scholar]
- 37.Schnitzler CE, et al. Genomic organization, evolution, and expression of photoprotein and opsin genes in Mnemiopsis leidyi: a new view of ctenophore photocytes. BMC Biol. 2012;10:107. doi: 10.1186/1741-7007-10-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Satterlie R, Case J. Gap junctions suggest epithelial conduction within the comb plates of the ctenophore Pleurobrachia bachei. Cell Tissue Res. 1978;193:87–91. doi: 10.1007/BF00221603. [DOI] [PubMed] [Google Scholar]
- 39.Steinmetz PRH, et al. Independent evolution of striated muscles in cnidarians and bilaterians. Nature. 2012;487:231–234. doi: 10.1038/nature11180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sebé-Pedrós A, et al. Insights into the Origin of Metazoan Filopodia and Microvilli. Mol Biol Evol. 2013;30:2013–2023. doi: 10.1093/molbev/mst110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tudor JE, Pallaghy PK, Pennington MW, Norton RS. Solution structure of ShK toxin, a novel potassium channel inhibitor from a sea anemone. Nat Struct Biol. 1996;3:317–20. doi: 10.1038/nsb0496-317. [DOI] [PubMed] [Google Scholar]
- 42.Marlow H, Arendt D. Evolution: Ctenophore Genomes and the Origin of Neurons. Curr Biol. 2014;24:R757–R761. doi: 10.1016/j.cub.2014.06.057. [DOI] [PubMed] [Google Scholar]
- 43.Smith CL, et al. Novel Cell Types, Neurosecretory Cells, and Body Plan of the Early-Diverging Metazoan Trichoplax adhaerens. Curr Biol. 2014;24:1565–1572. doi: 10.1016/j.cub.2014.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Senatore A, Reese TS, Smith CL. Neuropeptidergic integration of behavior in Trichoplax adhaerens, an animal without synapses. J Exp Biol. 2017;220:3381–3390. doi: 10.1242/jeb.162396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nikitin M. Bioinformatic prediction of Trichoplax adhaerens regulatory peptides. Gen Comp Endocrinol. 2015;212:145–155. doi: 10.1016/j.ygcen.2014.03.049. [DOI] [PubMed] [Google Scholar]
- 46.Riesgo A, Farrar N, Windsor PJ, Giribet G, Leys SP. The analysis of eight transcriptomes from all poriferan classes reveals surprising genetic complexity in sponges. Mol Biol Evol. 2014;31:1102–20. doi: 10.1093/molbev/msu057. [DOI] [PubMed] [Google Scholar]
- 47.Tautz D, Domazet-Lošo T. The evolutionary origin of orphan genes. Nat Rev Genet. 2011;12:692–702. doi: 10.1038/nrg3053. [DOI] [PubMed] [Google Scholar]
- 48.Sebé-Pedrós A, et al. High-Throughput Proteomics Reveals the Unicellular Roots of Animal Phosphosignaling and Cell Differentiation. Dev Cell. 2016;39:186–197. doi: 10.1016/j.devcel.2016.09.019. [DOI] [PubMed] [Google Scholar]
- 49.Levine M, Tjian R. Transcription regulation and animal diversity. Nature. 2003;424:147–51. doi: 10.1038/nature01763. [DOI] [PubMed] [Google Scholar]
- 50.Piasecki BP, Burghoorn J, Swoboda P. Regulatory Factor X (RFX)-mediated transcriptional rewiring of ciliary genes in animals. Proc Natl Acad Sci. 2010;107:12969–12974. doi: 10.1073/pnas.0914241107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang S, Samakovlis C. Grainy Head and Its Target Genes in Epithelial Morphogenesis and Wound Healing. Curr Top Dev Biol. 2012;98:35–63. doi: 10.1016/B978-0-12-386499-4.00002-1. [DOI] [PubMed] [Google Scholar]
- 52.Peter IS, Davidson EH. Evolution of gene regulatory networks controlling body plan development. Cell. 2011;144:970–85. doi: 10.1016/j.cell.2011.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sebé-Pedrós A, et al. The Dynamic Regulatory Genome of Capsaspora and the Origin of Animal Multicellularity. Cell. 2016;165:1224–1237. doi: 10.1016/j.cell.2016.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lara-Astiaso D, et al. Chromatin state dynamics during blood formation. Science. 2014;345:943–949. doi: 10.1126/science.1256271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nora EP, et al. Targeted Degradation of CTCF Decouples Local Insulation of Chromosome Domains from Genomic Compartmentalization. Cell. 2017;169:930–944.e22. doi: 10.1016/j.cell.2017.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang Q, Sun Q, Czajkowsky DM, Shao Z. Sub-kb Hi-C in D. melanogaster reveals conserved characteristics of TADs between insect and mammalian cells. Nat Commun. 2018;9:188. doi: 10.1038/s41467-017-02526-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ramírez F, et al. High-resolution TADs reveal DNA sequences underlying genome organization in flies. Nat Commun. 2018;9:189. doi: 10.1038/s41467-017-02525-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vij S, et al. Evolutionarily Ancient Association of the FoxJ1 Transcription Factor with the Motile Ciliogenic Program. PLoS Genet. 2012;8:e1003019. doi: 10.1371/journal.pgen.1003019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ganz T. Defensins: antimicrobial peptides of innate immunity. Nat Rev Immunol. 2003;3:710–720. doi: 10.1038/nri1180. [DOI] [PubMed] [Google Scholar]
- 60.Grell KG, Benwitz G. Ultrastruktur von Trichoplax adhaerens F.E. Schulze. Cytobiologie. 1971;4:216–240. [Google Scholar]
- 61.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Heinz S, et al. Simple Combinations of Lineage-Determining Transcription Factors Prime cis-Regulatory Elements Required for Macrophage and B Cell Identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jolma A, et al. DNA-binding specificities of human transcription factors. Cell. 2013;152:327–39. doi: 10.1016/j.cell.2012.12.009. [DOI] [PubMed] [Google Scholar]
- 65.Jolma A, et al. DNA-dependent formation of transcription factor pairs alters their binding specificity. Nature. 2015;527:384–388. doi: 10.1038/nature15518. [DOI] [PubMed] [Google Scholar]
- 66.Kulakovskiy IV, et al. HOCOMOCO: a comprehensive collection of human transcription factor binding sites models. Nucleic Acids Res. 2013;41:D195–202. doi: 10.1093/nar/gks1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tanay A. Extensive low-affinity transcriptional interactions in the yeast genome. Genome Res. 2006;16:962–972. doi: 10.1101/gr.5113606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Punta M, et al. The Pfam protein families database. Nucleic Acids Res. 2012;40:D290–301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.de Mendoza A, et al. Transcription factor evolution in eukaryotes and the assembly of the regulatory toolkit in multicellular lineages. Proc Natl Acad Sci. 2013;110:E4858–66. doi: 10.1073/pnas.1311818110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Katoh K, Toh H. Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform. 2008;9:286–298. doi: 10.1093/bib/bbn013. [DOI] [PubMed] [Google Scholar]
- 71.Darriba D, Taboada GL, Doallo R, Posada D. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics. 2011;27:1164–5. doi: 10.1093/bioinformatics/btr088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 73.Lartillot N, Lepage T, Blanquart S. PhyloBayes 3: a Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics. 2009;25:2286–2288. doi: 10.1093/bioinformatics/btp368. [DOI] [PubMed] [Google Scholar]
- 74.Li L, Stoeckert CJ, Jr, Roos DSD. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.