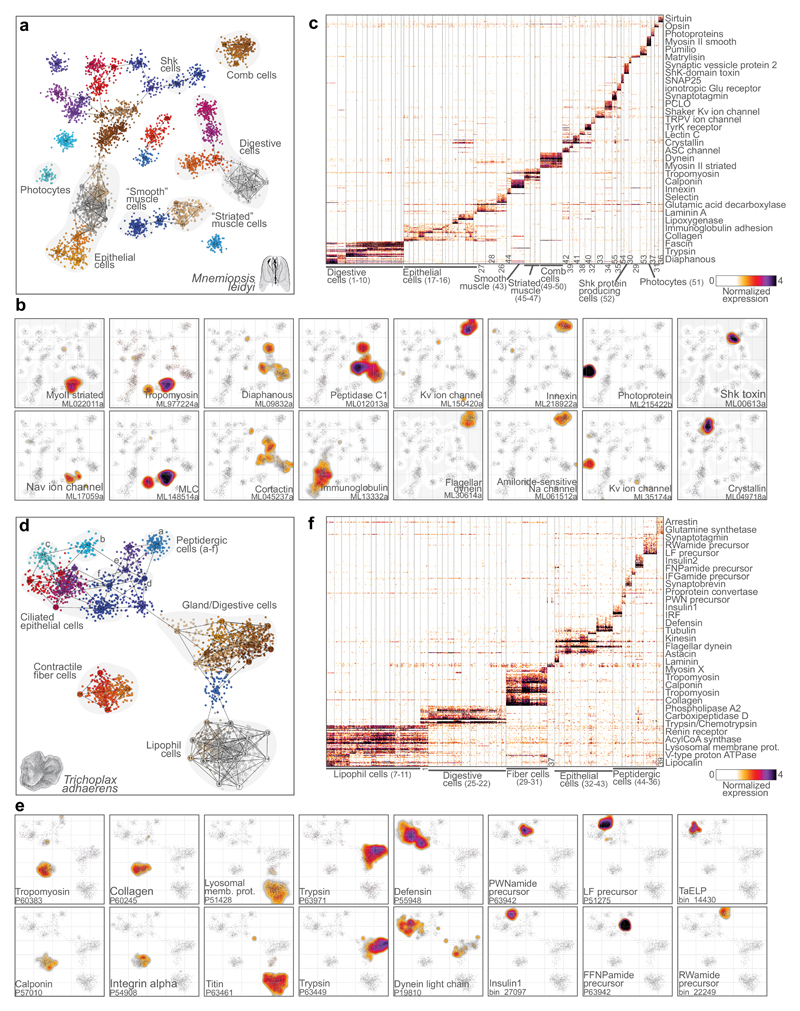
Figure 3. Mnemiopsis leidyi and Trichoplax adhaerens cell type atlases.
a, 2D projection of M.leidyi metacells and single cells. b, Gene expression distribution on 2D projected M.leidyi cells for selected gene markers. c, Normalized gene expression across 4,803 M.leidyi single cells (columns), sorted by cell cluster. For each cluster, the top 25 genes sorted by fold change versus the other metacells were selected for visualization (with a FC threshold>=2). d, 2D projection of T.adhaerens metacells and single cells. e, Gene expression distribution on 2D projected T.adhaerens cells for selected gene markers. f, Normalized gene expression across 3,209 T.adhaerens single cells (columns), sorted by cell cluster. Genes selected as in c. Color-coding of cells and metacells in a and d is arbitrary.