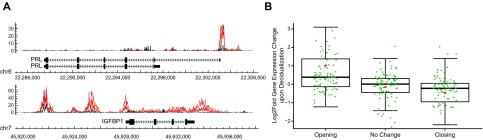
Figure 1.
ATAC-seq analysis of decidualizing EnSCs. A) Dynamic chromatin changes at key decidual marker genes in differentiating EnSCs. ATAC-seq signals in undifferentiated and decidualizing cells are shown in black and red, respectively. Proximal promoters of PRL (prolactin; upper panel) and IGFBP1 (IGF binding protein-1; lower panel) exhibit chromatin opening upon decidualization. IGFBP1 has additional regions of opening chromatin that are upstream of the TSS and downstream of the termination site. B) Dynamic chromatin changes in decidualizing EnSCs correlate with differential gene expression. Box plots show differential expression of 100 genes nearby (within 10 kb of TSS) the most opening or closing regions. Log2 fold change in expression is shown on the y axis: +ve and −ve values represent genes that are up- and down-regulated, respectively. ATAC-seq peaks were grouped according to whether they open, close, or remain unchanged upon decidualization. Red asterisk shows mean log2-fold change.