Figure 5 |. Chemical screening in the zebrafish.
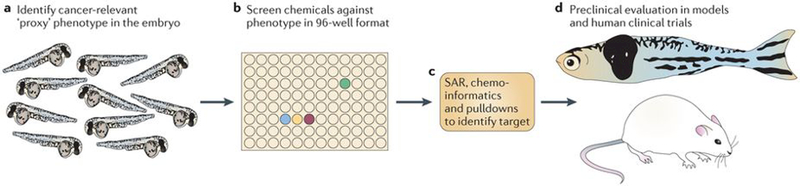
These types of screens are most efficiently carried out on embryos, given their amenability to large-scale, high-throughput manipulation and analysis. a | Identification of an embryonic phenotype that is highly relevant to cancer is a key step in this process. An embryo stained for the neural crest marker crestin is shown; neural crest cells give rise to pigmented melanocytes but also to melanoma in the zebrafish. b | After a relevant embryo phenotype is found, the embryos can be distributed in their chorions to plates, most typically the 96-well format. Each well will receive a distinct small molecule, either manually or with the aid of a liquid-handling robot. This method has been applied to screens ranging from 1,000 to 26,000 molecules. c | Identifying the mechanism of hits is shown. This will primarily depend on the nature of the library used. Molecules with unknown function may require methods such as structure–activity relationships (SARs), chemoinformatics (using algorithms and databases such as PubChem, ChemBank or DiscoveryGate) or pulldowns using tagged versions of drugs and mass spectrometry. For libraries biased towards chemicals with US Food and Drug Administration-approved or known mechanisms, this step can often be rapid, whereas for molecules of unknown function it can take up to 1 year or more. d | While mechanistic evaluation is ongoing, chemicals can be tested for their effects on cancer in multiple downstream assays, including zebrafish cancer models or mouse transgenic and xenograft models. Depending on potency and safety, some of these hits will be amenable to testing in clinical trials in humans.
