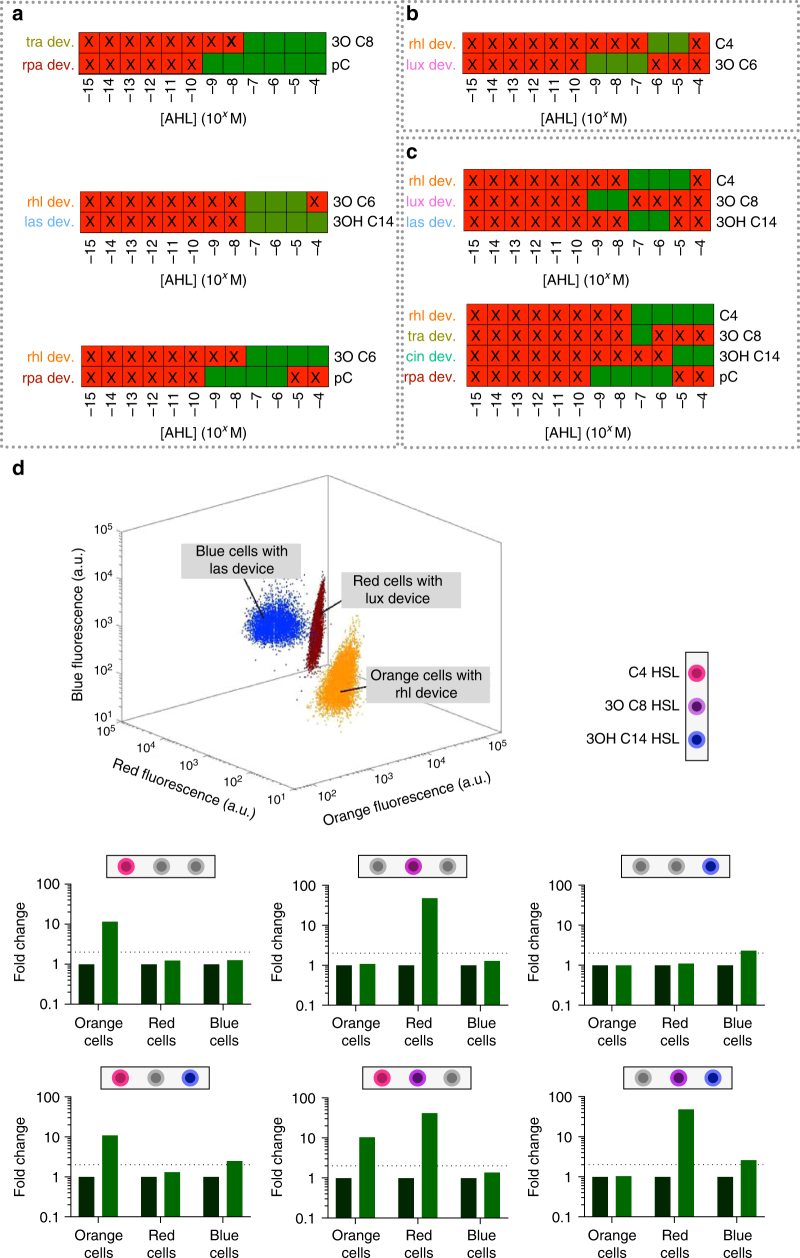
Fig. 4.
Computer-aided design of synthetic microbial consortia with orthogonal communication channels. a–c Orthogonal AHL-chemical communication channels as identified by our computer-aided-design software. For each panel, crossed red indicates an AHL inducer concentration which does not meet at least one of the user-defined specifications, while green indicates an AHL inducer concentration for which all user-defined specifications are met. a Systems of two communication channels designed with the following specifications: two-fold-specific gene activation and less than two-fold crosstalk. b System of two communication channels designed with the following specifications: more than ten-fold specific gene activation and less than three-fold crosstalk. c System of three communication channels (top) and four communication channels (bottom) designed with the following specifications: more than two-fold-specific gene activation and less than two-fold crosstalk. d Co-culture of three E. coli populations forming a system that is predicted to allow for orthogonal control of gene expression by the use of three chemical channels (as proposed in c: top illustration); Individual cell populations in co-culture were differentiated by flow cytometry by recording cell fluorescence resulting from mOrange (orange cell population), mRFP1 (red cell population) or mtagBFP2 (blue cell population) expression (3D-pot). The calculated fold change in GFP output for each cell population in response to chemical induction is indicated in the bar charts below the 3D-plot. The dotted line in the bar charts represents the user-specified threshold value for gene activation and crosstalk signal. Derived fold change values were calculated from one experimental implementation of the co-culture