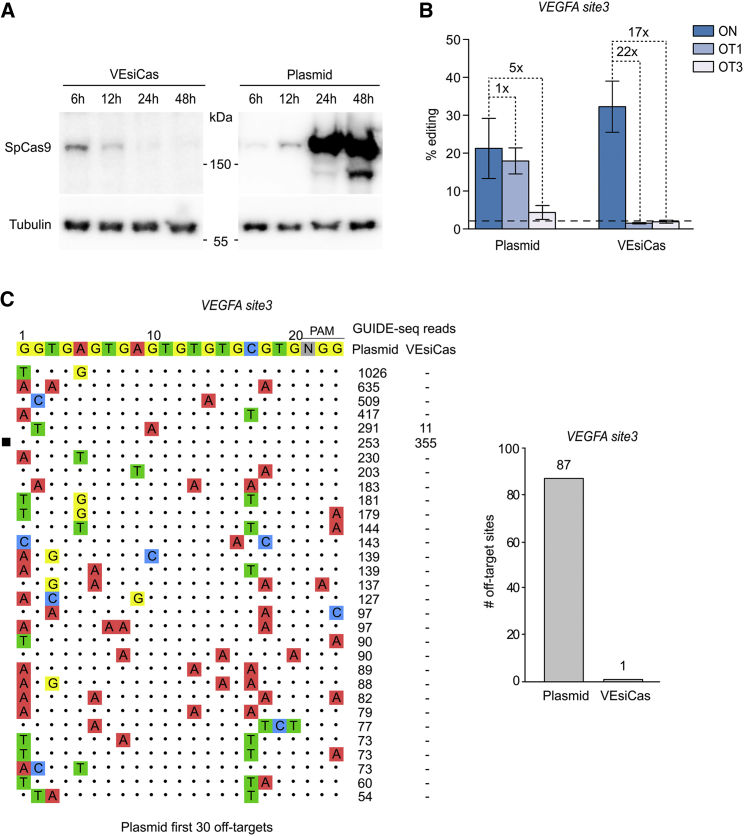
Figure 4.
Analysis of On/Off-Target Activity Generated by VEsiCas on the VEGFA Locus
(A) Traceless delivery of SpCas9 through VEsiCas. Shown is a time course analysis of SpCas9 intracellular levels by western blot analysis in HEK293T cells transduced with VEsiCas or transfected with a plasmid expressing SpCas9. The reported time points correspond to the time of analysis following treatments (transduction or transfection). The western blot is representative of n = 2 independent experiments. (B) Targeted comparison of genome editing specificity using VEsiCas or a plasmid expressing SpCas9 in combination with sgVEGFA site3. Shown are percentages of indel formation measured by TIDE in the VEGFA site3 locus and in two sgVEGFA site3 previously validated off-target sites (OT1 and OT3) in HEK293T cells. The dashed bars represent the TIDE background. (C) Genome-wide specificity of VEsiCas targeting the VEGFA site3 locus. Shown is GUIDE-seq analysis for the sgRNA targeting VEGFA site3 locus in HEK293T cells transfected with SpCas9 or treated with VEsiCas. The black square indicates on-target sites. DNA from three biological replicates was mixed before library preparation. Right: the total number of off-targets identified for the sgVEGFA site3 with SpCas9 transfection or VEsiCas treatments.