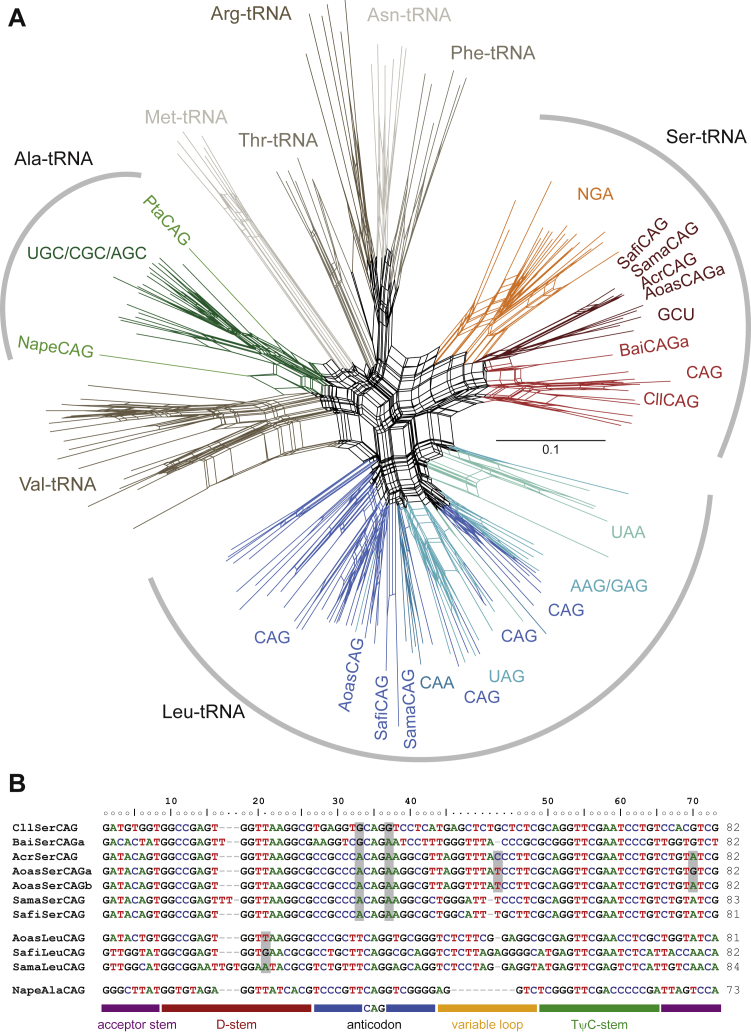
Figure 2.
Yeast tRNACAG Phylogeny and Sequences
(A) An unrooted phylogenetic network of Leu-, Ser-, and Ala-tRNAs was generated with SplitsTree. Selections of Val-, Met-, Thr-, Arg-, Asn-, and Phe-tRNAs were added as outgroups. Leu-, Ser-, and Ala-tRNAs are colored by isoacceptor. CUG-encoding tRNAs of P. tannophilus (Pta), N. peltata (Nape), A. rubescens (Acr), A. asiatica (Aoas), S. fibuligera (Safi), S. malanga (Sama), B. inositovora (Bai), and C. lusitaniae (Cll) are highlighted for better orientation. Identical tRNA identity assignments are obtained in phylogenetic analyses using maximum-likelihood and Bayesian approaches.
(B) Sequences of the tRNACAG of the analyzed species. Some nucleotides are highlighted for faster orientation. The Saccharomycopsis differ most strikingly from the A. asiatica at the important position 20a, at which the presence of purine nucleotides has been shown to dramatically reduce leucylation efficiency. Guanine nucleotides 5′ and 3′ of the anticodon have been shown to cause low-level misleucylation in vitro, but the whole-cell proteomics data of C. lusitaniae show unambiguous CUG translation as serine in vivo. The two A. asiatica differ from the A. rubescens by only 1 and 2 nt and both substitutions are at variable positions.