Figure 5.
Yeast Phylogeny
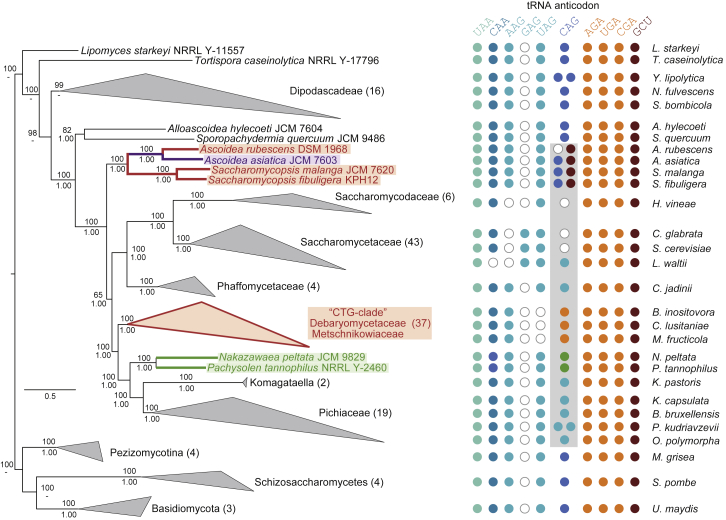
RAxML-generated phylogeny of 137 yeast species and 11 fungal outgroup species. Support values for major branches are given as bootstrapping replicates (RAxML-generated tree, numbers above branches) and posterior probabilities (MrBayes-generated tree, numbers below branches). For display purposes, species of major lineages have been collapsed into groups, with the numbers in parentheses denoting the number of species and the corners of the triangles representing the shortest and longest distances within the groups. Species and groups employing alternative genetic codes are indicated by color: “CTG clade” (DM clade) species A. rubescens, S. malanga, and S. fibuligera encode CUG as serine (red), N. peltata and P. tannophilus as alanine (green), and A. asiatica encodes CUG as both leucine and serine (purple). The grouping of the Ascoidea yeasts is consistent with a recent phylogenomics study [15]. Discrepancies with other published trees, which placed Ascoidea sister to the Phaffomycetaceae/Saccharomycetaceae/Saccharomycodaceae [34] or the Pichiaceae [17], are best explained by the deeper taxonomic sampling of early-branching yeasts (24 versus 10) in our study and the considerably increased sequence data (26 proteins versus 5), respectively. For each yeast, the presence (colored dots) and absence (white dots) of tRNA isoacceptors encoding the leucine (blue colors) and serine (red colors) codons are depicted at the side of the tree. For lineages collapsed in the tree, the tRNA repertoire of representative species is given. See also Figures S5 and S6.