Fig. 2.
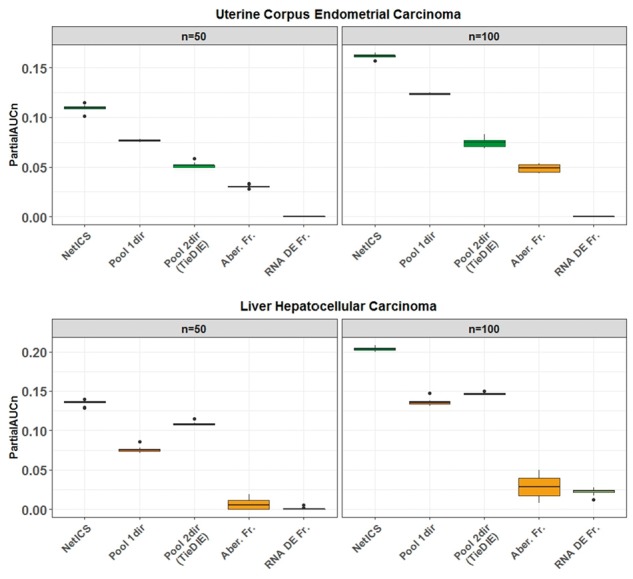
Comparison of gene prioritization methods. We compared the performance of NetICS to four methods including pooling aberrant genes from all samples before diffusion (Pool1dir), pooling both aberrant and differentially expressed genes from all samples before bidirectional diffusion (Pool2dir), ranking by frequency of aberrant genes across all samples (Aber. Fr.) and ranking by frequency of differentially expressed genes across all samples (RNA DE Fr.). By bootstrapping the available samples 10 times, we computed the partial AUC for n = 50, 100 (x-axis). The performance was tested on the TCGA datasets of uterine corpus endometrial carcinoma (top) and liver hepatocellular carcinoma (bottom)
