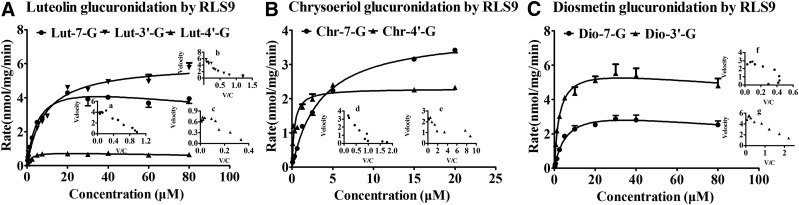
Fig. 4.
Kinetics of luteolin (A), chrysoeriol (B), and diosmetin (C) glucuronidation by RLS9 at different concentrations (0.3125–80 μM for luteolin and diosmetin; 0.0781–20 μM for chrysoeriol). Each inset shows the Eadie–Hofstee plot. For luteolin glucuronidation, the formations of Lut-7-G (Aa) and Lut-4′-G (Ac) were fitted using the substrate inhibition model; Lut-3′-G (Bb) formation followed the Michaelis–Menten equation. For chrysoeriol glucuronidation, the formations of Chr-7-G (Bd) and Chr-4′-G (Be) were fitted using the Michaelis–Menten equation. For diosmetin glucuronidation, Dio-7-G (Cf) and Dio-3′-G (Cg) formation followed the autoactivation and the substrate inhibition model, respectively. The RLS9 concentrations were 0.005–0.01 mg/mL, and the incubation time was 30 minutes. All incubations were performed in triplicate. Each point represents the mean of three determinations. The error bar represents the S.D. of the mean (n = 3, mean ± S.D.).