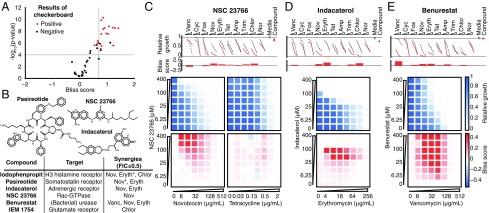
Fig. 4.
Validation of hits from primary screen. (A) In eight-point checkerboard assays with E. coli, we tested a total of 46 compound × antibiotic combinations, of which 17 (11 distinct compounds) scored as hits in the primary screen (SI Appendix, Figs. S17 and S18 and Dataset S4). Combinations that scored positive (red) and negative (black) for Bliss synergy are plotted according to results from the primary screen. Gray dotted lines indicate primary screening thresholds. (B) Target, status, antibiotic synergy set (by FIC index method), and selected structures of validated hits (SI Appendix, Figs. S17, S19, and S20 and Dataset S3). The first four compounds are from the full-scale phase; the last two are from the pilot phase. Antibiotics marked with an asterisk represent additional synergies revealed in validation. Structures were rendered in ChemDraw from SMILES strings. (C, Top) Primary screening data and calculated Bliss scores for NSC 23766. Growth in presence of compounds alone (Compound, indicated by red dots) relative to the absence of antibiotic and compound (Media, indicated by gray dots) is shown. (C, Bottom) The 96-well plate checkerboard assay of NSC 23766 × novobiocin (positive for synergy) (Left) and NSC 23766 × tetracycline (negative for synergy) (Right). For each checkerboard assay in C–E, the relative growth values and Bliss scores are shown (color scaling is indicated at Far Right). (D) Primary screening and checkerboard data for indacaterol × erythromycin (positive for synergy). (E) Primary screening and checkerboard data for benurestat × vancomycin (positive for synergy). Amp, ampicillin; Chlor, chloramphenicol; Cyc, cycloserine; Eryth, erythromycin; Fos, fosfomycin; Nor, norfloxacin; Nov, novobiocin; Tet, tetracycline; Trim, trimethoprim; Vanc, vancomycin.