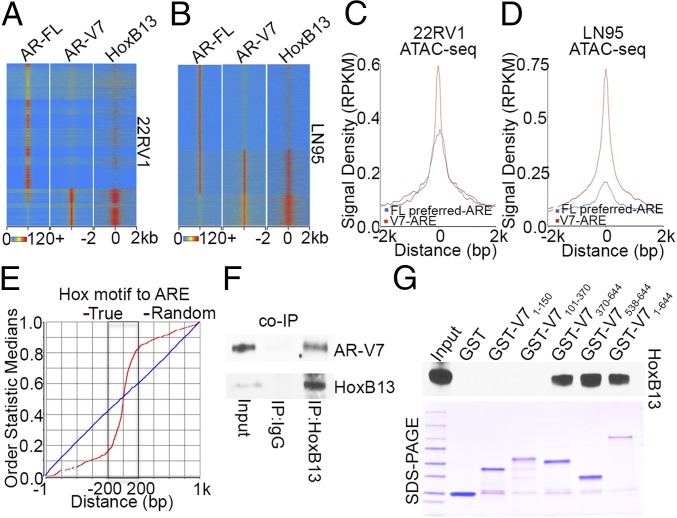
Fig. 2.
AR-V7 and HoxB13 interact genomically and physically. (A and B) Heatmaps show the ChIP-exo signal intensity of AR-FL, AR-V7, and HoxB13 binding in 22RV1 cells (A) and LN95 cells (B). (C and D) Chromatin accessibility in 22RV1 (C) and LN95 (D) cells was determined by ATAC-seq. Normalized averaged ATAC-seq signal tag distribution over AR-V7 ARE regions and AR-FL preferred-ARE regions is shown. The window indicates ±2-kb regions of AREs. (E) Cumulative genomic position distributions of precisely defined HoxB13 motifs relative to AR-V7–bound AREs in AR-V7/HoxB13 binding locations in 22RV1 and LN95 cells. (F) Whole-cell lysates from hormone-depleted 22RV1 cells were immunoprecipitated with HoxB13 antibodies, and Western blots were performed with antibodies against AR-V7 and HoxB13. (G) GST pull-down assays were performed by incubating in vitro translated HoxB13 with GST ∼ AR-V7 fusion proteins. Western blots were performed using an anti-HoxB13 antibody.