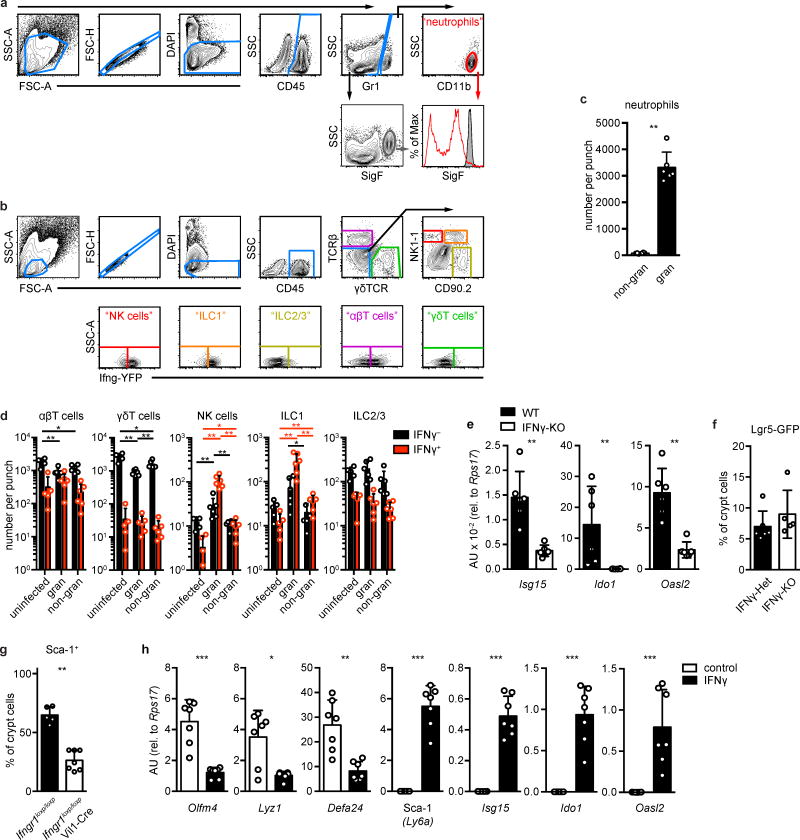
Extended Data Figure 4. IFNγ produced by Hp-responsive immune cells drives the granuloma gene signature.
a–f, Mice were infected with H. polygyrus (Hp) and analyzed at day 6, unless otherwise indicated. a–b, Representative gating example of neutrophils (a) and NK cells, ILC1, ILC2/3, αβ T cells, and γδ T cells (b). c, Neutrophils were enumerated from non-granuloma (“non-gran”) or granuloma (“gran”) biopsies. n=6. d, Ifng reporter mice were untreated (uninfected) or infected (gran, non-gran) with Hp and analyzed 5–6 days later for hematopoietic (CD45+) populations: NK cells, ILC1, ILC2/3, αβ T cells, γδ T cells. No reporter signal was seen in non-lymphoid populations. n=5 (uninfected) or 6 (gran, non-gran). e, Crypt cells were sorted from granuloma biopsies of IFNγ-null (KO) mice and analyzed for the indicated transcripts. n=6/group. f, Lgr5-GFP mice were bred to IFNγ-KO mice and analyzed for Lgr5-GFP expression in crypt epithelia from granuloma biopsies. n=6 (Het) or 5 (KO). g, Ifngr1loxp/loxp mice were bred to Vil1-Cre mice and analyzed for Sca-1 expression in crypt epithelia from granuloma biopsies. n=5 (Ifngr1loxp/loxp) or 7 (Ifngr1loxp/loxp;Vil1-Cre). h, Wild type organoids were treated with 5 ng/ml IFNγ for 24 hours and analyzed for the indicated transcripts. n=7 cultures per group. Unpaired, two-tailed Mann-Whitney test; mean ± S.D. (c–h). * P < 0.05, ** P < 0.01, *** P < 0.001.