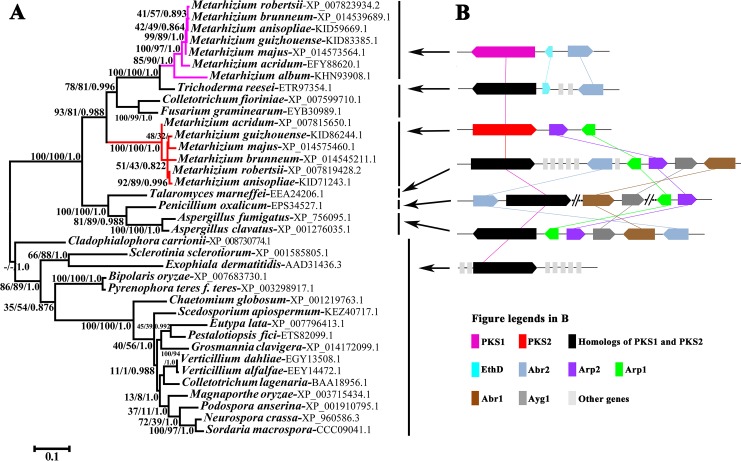
Fig 1. Phylogenetic and syntenic assays of PKS1 and PKS2 from M. robertsii, and their homologs in other ascomycete fungi.
(A) Phylogenetic tree based on the amino acid sequences of the PKS KS domains. The Genbank accession number for each full-length amino acid sequence of each PKS is shown after the species name. The clade containing the Metarhizium PKS1s is highlighted in pink, that containing PKS2s is highlighted in red. Numbers at nodes represent the bootstrap values of Maximum Likelihood (left), Neighbor-Joining (middle), and the Bayesian posterior probabilities (right). A hyphen (-) indicates no support for the given node in the corresponding method. The scale bar corresponds to the estimated number of amino acid substitutions per site. (B) Schematic maps of the gene clusters containing Pks1, Pks2, or their homologs. Colors in the inset legend represent genes in the cluster; gray bars indicate genes that are not homologous to each other. The groups of fungal species that contain specific gene clusters are indicated with arrows.