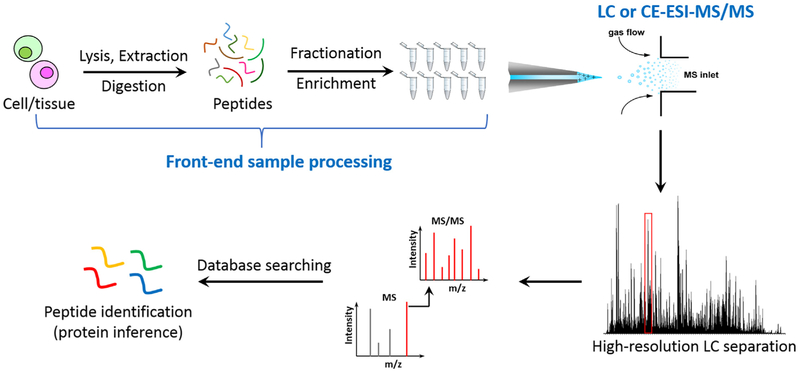
Fig. 2.
A general workflow of bottom-up MS-based proteomics. It typically starts from cell lysis, protein extraction, and proteolytic digestion to peptide mixtures, followed up by fractionation or enrichment procedures and LC or CE-MS/MS analyses. The experimental spectrum is matched with database to identify peptides or contaminants.