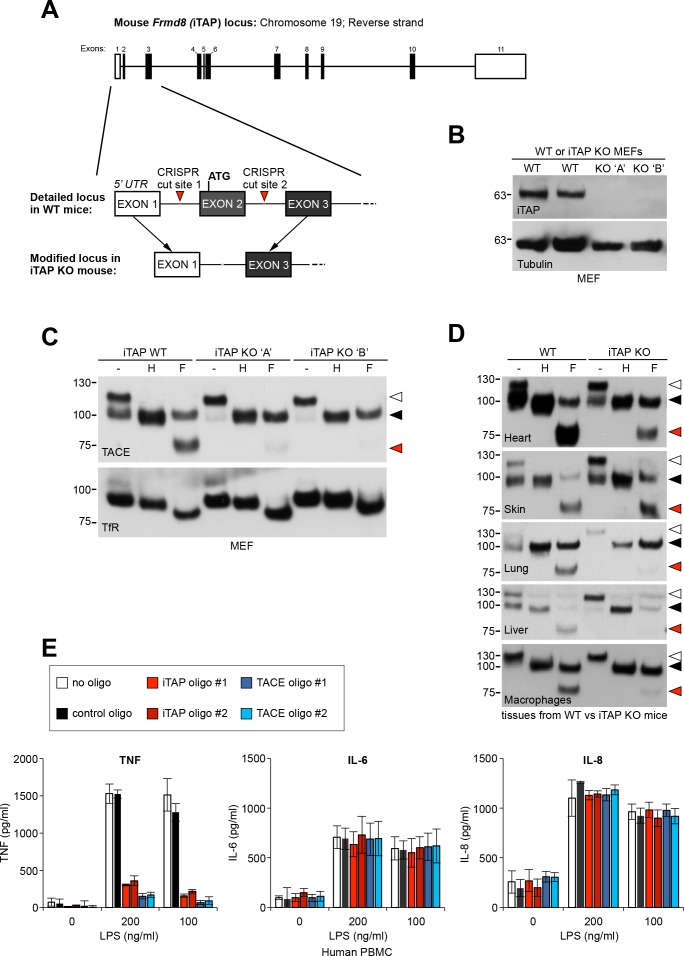
Figure 7. iTAP is essential for TACE maturation and function in primary cells and tissues from human and mouse.
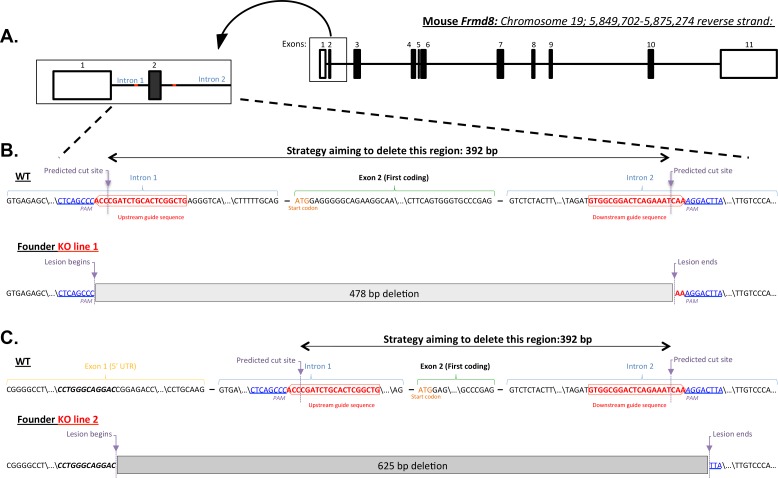
(A). Schematic representation of the CRISPR targeting strategy to delete mouse Frmd8 (iTAP) gene using two guide RNAs flanking the first coding exon (exon 2). In the upper schematic of the Frmd8 locus, open boxes indicate non-coding exons whereas filled boxes indicate coding exons (B). Mouse embryonic fibroblasts (MEFs) were isolated from WT versus two independent iTAP KO E14.5 embryo littermates. The loss of iTAP at the protein level is shown by immunoblotting. (C). Mature TACE is diminished in iTAP KO MEFs. ConA-enriched lysates from MEFs isolated from WT versus iTAP KO embryos were deglycosylated as described previously. The transferrin receptor (TfR) is used as a loading control. (D). Mature TACE is depleted or diminished in TACE-relevant tissues from iTAP KO mice. ConA-enriched lysates from WT vs iTAP KO mouse tissues and bone marrow-derived macrophages, were deglycosylated as described previously. TACE was detected by western blot. The immature and mature species of TACE are indicated with white arrowheads and black arrowheads respectively, whereas red arrowheads denote the fully deglycosylated mature polypeptide. The experiment was performed twice with lysates isolated from tissues from two individual KO mice. (E). iTAP is essential for TACE physiological regulation in human primary cells. Isolated primary human peripheral blood mononuclear cells (PBMC) were differentiated into monocytes, then electroporated with the indicated siRNAs. Cells were then stimulated with the indicated concentrations of lipopolysaccharide (LPS). After 18 hr, the concentration of the cytokines TNF, IL-6 and IL-8 secreted into the supernatants, was measured by ELISA. The experiment was done three independent times and data from one representative experiment is shown. Data presented as mean ± standard error from triplicate measurements.