Fig. 2.
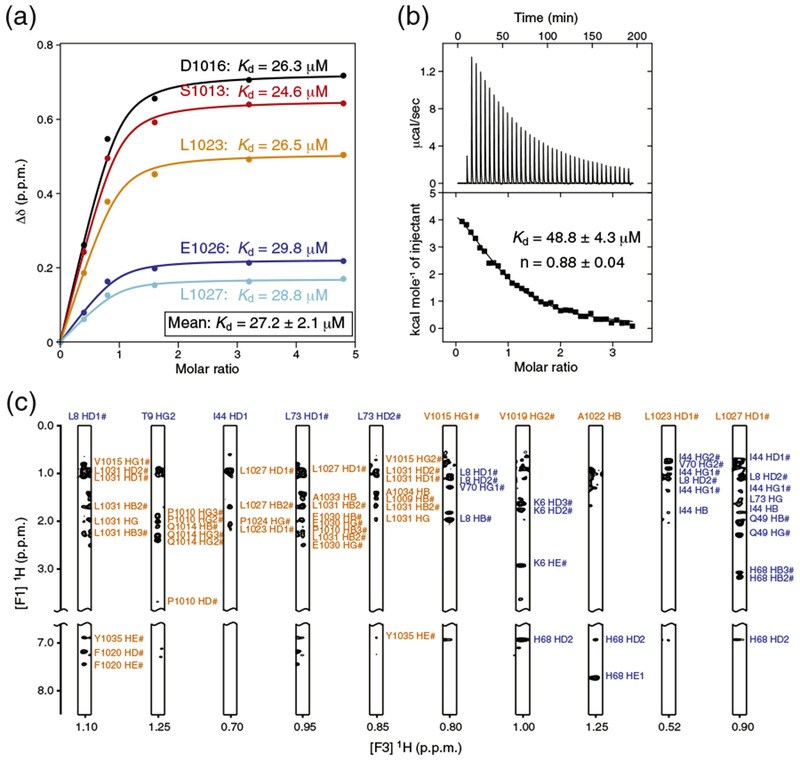
Estimation of the binding affinity of human REV1 (932–1039) for ubiquitin. (a) NMR spectroscopy-based titration of 15N-labeled REV1 with non-labeled ubiquitin. REV1 was at initial concentration of 0.5 mM. Multiple apparent Kd values were determined from the changes in chemical shifts for five REV1 residues by nonlinear least-squares analysis using equation: where [L] is the concentration of ubiquitin, [P] is the concentration of REV1, Δδ is the observed normalized chemical shift change as defined in Fig. 1b, and Δδmax is the normalized chemical shift change at saturation. (b) ITC of REV1 with ubiquitin. The integrated heat measurements from raw titration data and curve fitting using a one-site binding model are shown. The Kd and stoichiometry (n) values are indicated with the standard deviations determined by nonlinear least-squares fitting. (c) Representative intermolecular NOE signals identified in F1 13C-filtered, F2 13C-edited NOESY-HSQC spectra. The resonance labels are colored orange for REV1 and blue for ubiquitin.
