Fig. 3.
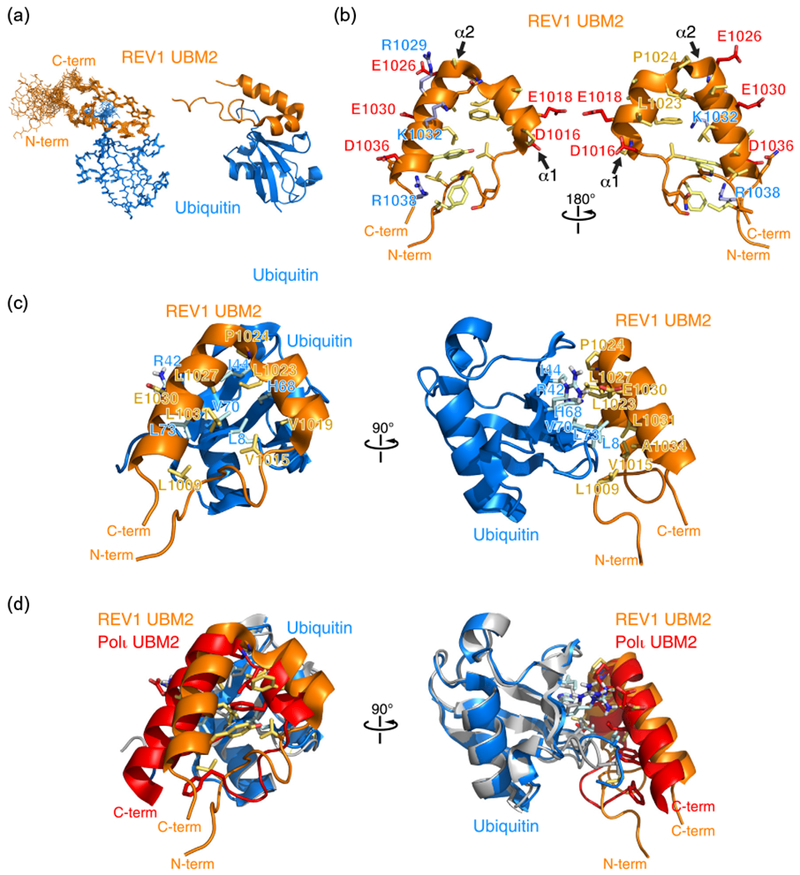
Three-dimensional structure of human REV1 (932–1039) in complex with ubiquitin. (a) Cartoon representation of the overlay of 20 REV1–ubiquitin complex structures representing the NMR ensemble (left) and a cartoon representation (right) of REV1 UBM2 in complex with ubiquitin. While structure calculations were done with REV1 (932–1039), the N-terminal disordered region is not shown. Only REV1 (1000–1039) is shown. (b) Cartoon and stick representation of the UBM2 structure highlighting negatively charged (blue), positively charged (red), neutral (orange), and hydrophobic (yellow) amino acid side chains. (c) Cartoon and stick representation of the REV1 UBM2–ubiquitin complex. Selected amino acid side chains at the binding interface are shown and labeled. (d) Overlay of the ubiquitin structures in the REV1 UBM2–ubiquitin and Polι UBM2–ubiquitin (PDB accession number 2KTF) complexes. Ubiquitin bound to REV1 is shown in blue, while ubiquitin bound to Polι is shown in gray.
