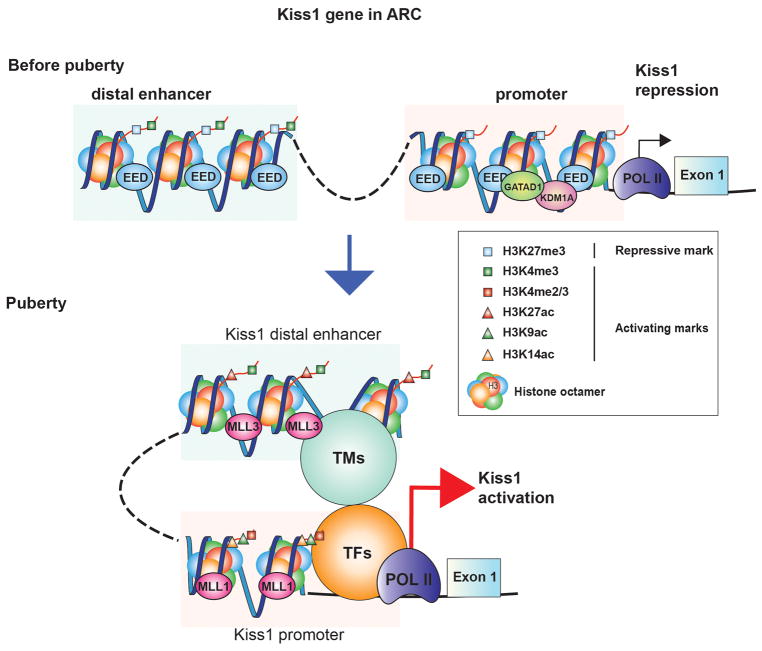
Figure 1. The epigenetic control of Kiss1 expression in the ARC.
This model postulates that during prepubertal development the transcriptional activity of the Kiss1 gene is repressed by a series of repressors, exemplified by the PcG as well as the GATAD1/KDM1A complex. By depositing histone PTMs associated with gene silencing (e.g. H3K27me3, blue squares) at Kiss1 promoter and enhancer sites, PcG proteins (epitomized by EED) act as ‘writers’ of a repressive chromatin configuration. Furthermore, the GATAD1 ‘reader’ protein recruits KDM1A, the ‘eraser’ of H3K4me2 further repressing chromatin accessibility. As puberty advances, these ‘writers’ and ‘readers’ are removed from these regulatory regions, and as a consequence of this loss, the amount of H3K27me3 is reduced, while H3K4me2/3 increases. As this transformation takes place, another set of writers, typified by the TrxG-activating multiprotein complex is engaged causing the deposition of histone PTMs linked to transcriptional activation at two sites: a) H3K4me2/3 (red squares) and H3K9,14ac (green and orange triangles) are added at promoter regions and, b) H3K4me3 and H3K27ac (red triangles) at enhancer sites characterized by their high and sustained content in H3K4me1 (green squares). This chromatin looping also brings a series of unknown transcription factors (TFs) like pioneer, determining and collaborative TFs, as well as transcriptional mediators (TMs) like Histone Acetyltransferases, DNA demethylation enzymes, Chromosomal looping factors, RNA processing factors, non-coding RNAs, etc [153]. An outcome of these changes is that the activated enhancer is attracted to the promoter region enhancing polymerase II (POLII) transcriptional activity, thus increasing Kiss1 mRNA transcription.