Figure 3. Overview of DPYD-Varifier.
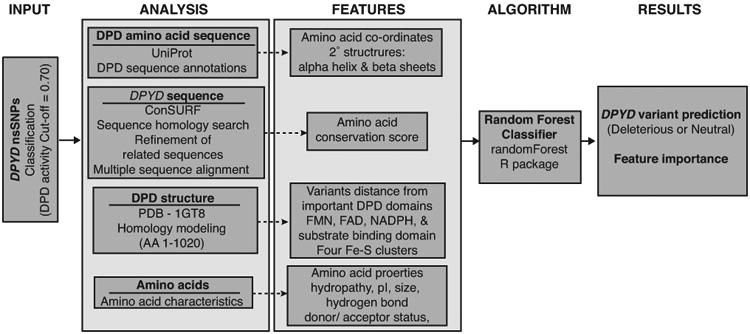
DPYD-Varifier was trained using relative DPD activity data obtained from in vitro experiments. DPYD and DPD sequence analysis was performed to obtain different features built on the classifier, including conservation scores, protein secondary structures, distance from domains, and amino acid biochemical properties. A random forest algorithm was utilized to obtain variant predictions and to assess feature importance to the model.
