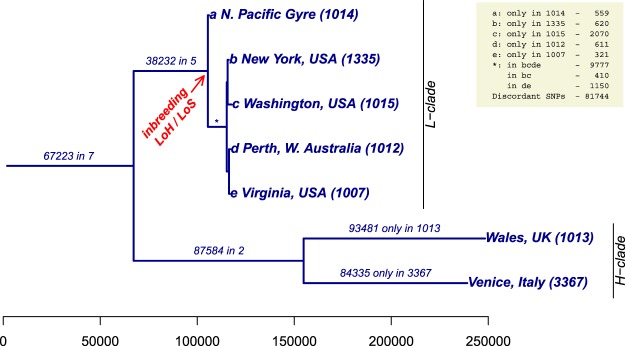
Figure 3.
SNP sharing among the seven strains of T. pseudonana. Cladogram based on a total of 468,117 refined SNPs, accounting for 82% of SNP positions, identified across all 7 strains (Supp. 9). The tree is rooted at the common ancestor of all seven isolates, with each split selected to maximize the number of SNPs shared by isolates within each subtree and minimize sharing across subtrees. Internal branch lengths are the numbers of SNPs shared by all isolates in the subtree below that branch. Terminal branch lengths are the numbers of private SNPs found in each isolate. The red arrow reflects an inferred evolutionary event in which inbreeding resulted in a significant loss of heterozygosity (LoH) and concomitant loss of sexual reproduction (LoS) that founded the L-clade. Scale: number of SNPs.