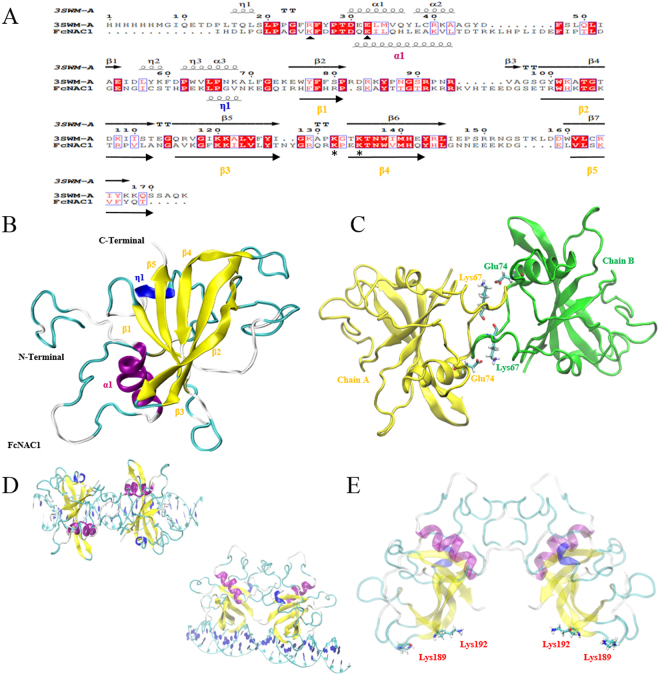
Figure 7.
Homology modeling of the NAC domain present in FcNAC1 protein. (A) The alignment between the template (3SWM chain A) and FcNAC1 NAC domain, including structural information. Spirals and arrows at the top of the sequences represent the secondary structure of the template, while those at the botton correspond to predicted secondary structure of FcNAC1. Triangles indicate the important residues for the dimerization process and asterisk indicate conserved residues for DNA interaction. (B) Homology modeling of FcNAC1 NAC domain using MODELLER software. Fifty models were built and the best was subjected to energy minimization and molecular dynamics with the purpose of relaxing the structure and minimizing steric impediments. It is possible to appreciate that the cluster of β sheets is flanked on both sides by the α-helix and the α-helix 310. α-helices were labeled as α, and β-sheets as β. (C) Homodimer structure of FcNAC1 NAC domain showing the potential residues involved in the salt/bridge interaction. (D) Protein-DNA positioning between the homodimer of FcNAC1 and the DNA sequence co- crystallized with the template 3SWM (5′- GTCTTGCGTGTTGGAACACGCAACAG -3′). (E) Visualization of the homodimer structure of FcNAC1 NAC domain highlighting the positively charged residues involved in the protein-DNA interaction.