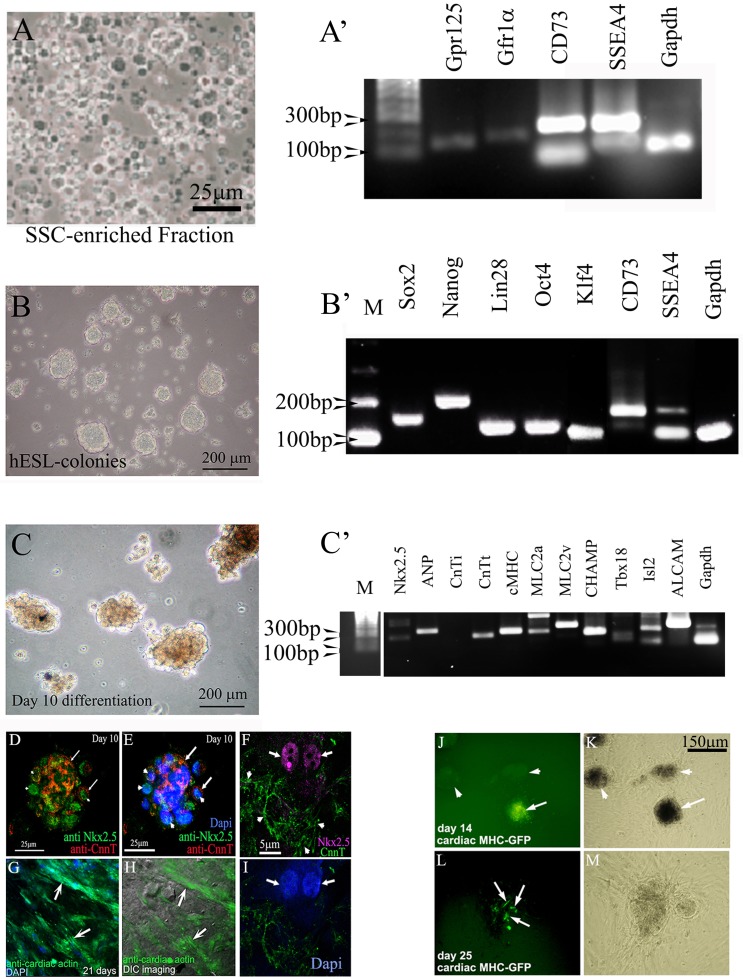
Figure 2.
Cardiac lineages can be produced from hgPSCs. A). Cells of the SSC-enriched fraction are cultured in medium containing GDNF for four days. The fraction is full of cells positive for various SSC markers including SSEA4. B) After four days, the medium is switched to a basic hESC medium containing bFGF and serum replacement. Cells are incubated for at least 10 days, after which rt-PCR shows evidence of all four Yamanaka factors plus nanog and CD73. C) Switching hESC medium for cardiac differentiation medium results in growth and morphologically darker looking colonies. RT-PCR shows expression of 9 out of 10 cardiac genes within 10 days of differentiation. D-I) Confocal analyses show protein expression of specific cardiac genes including nuclear staining of Nkx2.5 (F arrows). Arrows in D and E point to areas positive for cardiac troponin (cTNT), while arrowheads point to nuclear Nkx2.5 staining. Dapi staining in E identifies the nuclei. Arrows in G and H point to distinct filaments of cardiac actin. The DIC image in H reveals the actin fibers within healthy cells. J-M) Transfection of colonies with a cMHC-GFP further confirms cardiac gene expression. Arrow in J points to a GFP positive colony. Arrowheads point to untransfected colonies. These untransfected colonies serve as an internal control ruling out autofluorescence. L) Arrows point to cells within the colony expressing cMHC-GFP. K and M) Phase contrast views show healthy colonies.