Figure 1. DNA2 binds preferentially to centromeric DNA and is required for centromeric DNA replication.
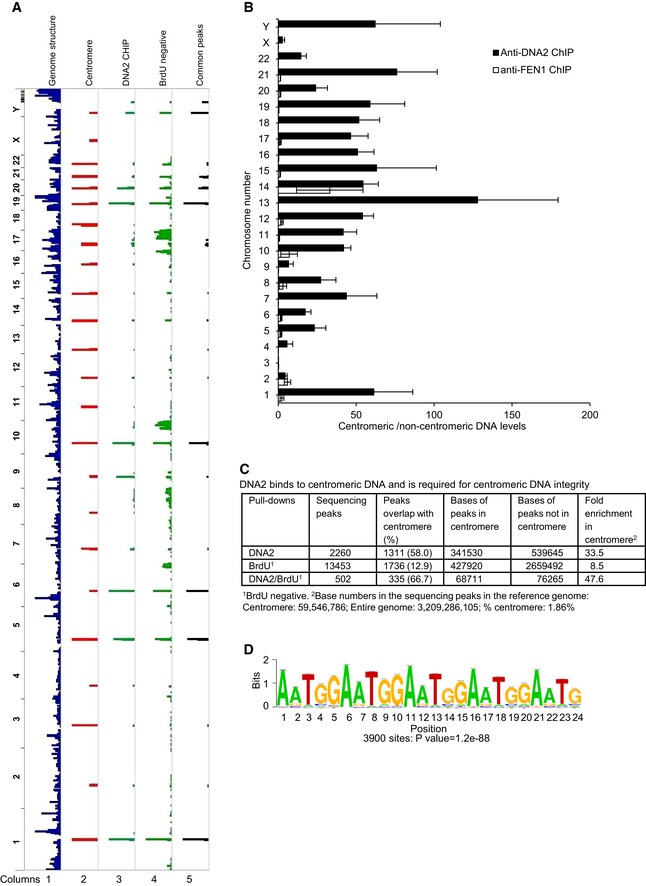
- DNA peaks were identified as described in Materials and Methods. Shown is an alignment of whole‐genome sequencing results to the Hg38 genome. Column 1, Hg38 genome structure (UCSC genome browser); column 2, Hg38 centromere locations; column 3, DNA2 binding sites in HCT‐116 cells identified from ChIP‐seq; column 4, BrdU‐negative DNA from DNA2‐null cells; column 5, common peaks among DNA samples shown in lanes 3 and 4. The chromosome number is listed on the left of the plot.
- Fold enrichment of centromeric versus non‐centromeric DNA over input as determined by qPCR of DNA2 and FEN1 ChIP samples. Shown by chromosome (Chr) are the means ± SDs of three biological repeats. See Table EV1 for primers used.
- Quantitative analysis of the DNA peaks and bases shown in panel (A).
- The most highly enriched repetitive centromeric/paracentromeric motif is shown. Letter size reflects its frequency at the position. P‐value of 1.2e‐88 derived by Fisher's exact test.
