Figure 3. Structure of the LUBAC Ternary Complex Core.
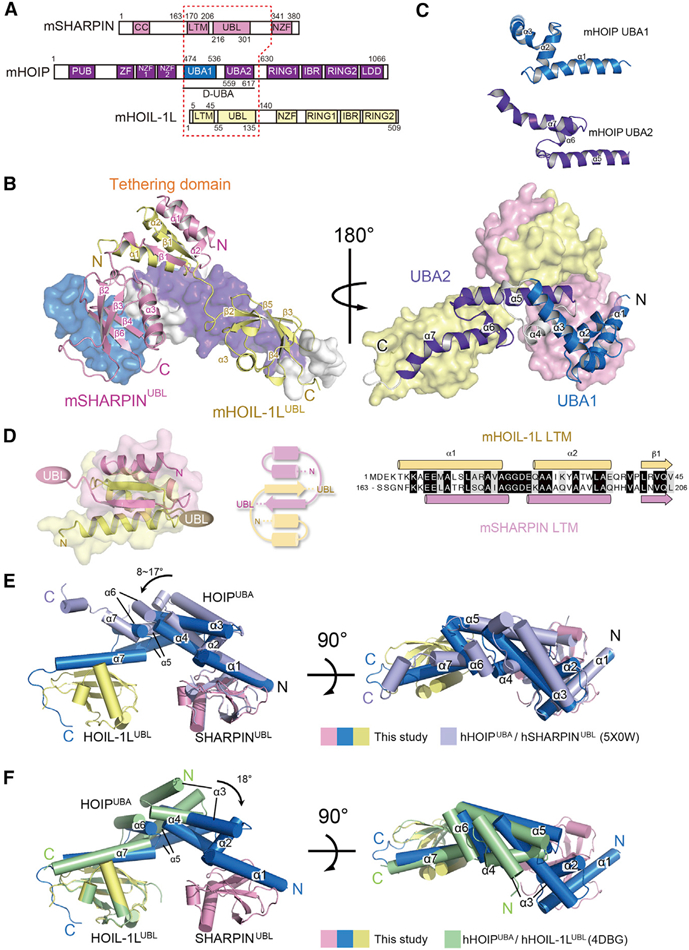
(A) Schematic representation of the domains of HOIL-1L, HOIP, and SHARPIN. Fragments of mouse subunits used for structural analysis are indicated by a red dotted frame.
(B) Overall structure ofthe LUBACternary complex core. The LUBAC core comprises mHOIP D-UBA (aa 474–630) (UBA1, cyan; UBA2, purple; and α4 helix and C-terminal loop, gray), HOIL-1L LTM-UBL(aa1–140) (yellow), and mSHARPIN LTM-UBL (aa 163–341) (pink). Structures are shown from two different viewpoints.
(C) Structural comparison between UBA1 and UBA2 ofmHOIP. Helices α–3 and α5–7arefolded into a common UBA module with atypical extended helices α1 and α7.
(D) Structure and topological diagram of TD. Sequence alignment of LTMs of mHOIL-1L and mSHARPIN are shown in the right panel. Identical and similar residues are highlighted in black and gray, respectively.
(E) Structural comparison between the ternary and hHOIP UBA/hSHARPIN UBL dimeric complexes. The structure of the hHOIP UBA/hSHARPIN UBL complex (PDB: 5X0W) is superposed onto that of the ternary complex core, based on the position of the UBLs ofSHARPIN. The structure ofthe ternary complex core is shown in the same color scheme used in Figure 3A. The representative hHOIP/hSHARPIN binary complex is presented in light purple.
(F) Structural comparison between the ternary and hHOIP UBA/hHOIL-1L UBL dimeric complexes. The structure of hHOIP UBA/hHOIL-1L UBL complex (PDB:4DBG) is superimposed ontothat ofthe ternary complex core, based on the UBLs. The structure of the hHOIP/hHOIL-1L complex is shown in green.
See also Figures S3 and S4 and Table S2.
