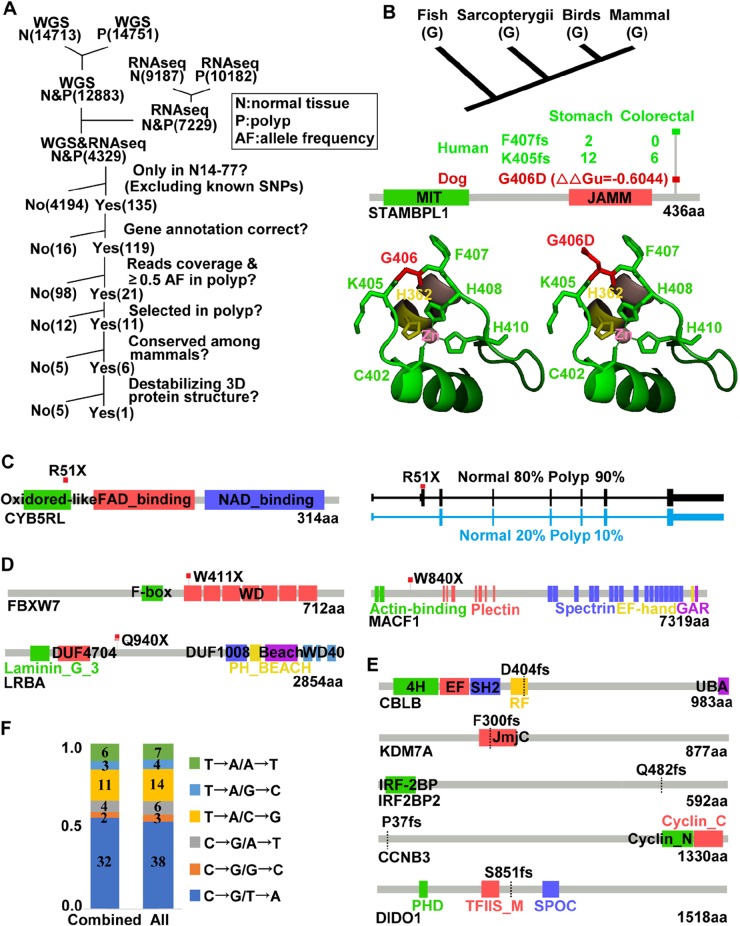
Figure 3. Notable germline and somatic mutations were identified in N14-77.
(A) Outlined is our pipeline for putative pathogenic germline missense mutation discovery (see text). Numbers in parentheses indicate mutation counts. (B) G406D of STAMBPL1 is identified by the pipeline in A. The top image shows G406 conservation. Middle images (from bottom to top) indicate the protein domains, ∆∆Gu (red) from modeling [24] predicting that G406D likely destabilizes the protein structure, and frameshift (fs) mutations found in human cancers (with total case numbers indicated). Bottom images are the crystal structure [25] with G406 (left) and G406D (right) in red and its flanking K405 and F407 in green, and zinc-coordinating residues C402, H362, H408 and H410 shown. (C) R51X of CYB5RL is a heterozygous germline truncation mutation. The right image indicates the two alternative splicing forms and their proportions in each sample. (D and E) Somatic truncation mutations and frameshift mutations identified. (F) C→T/G→A changes dominate. Combined: somatic missense mutations identified by combining WGS and RNA-seq reads. All: those found by WGS alone, RNA-seq alone and combined. The numbers inside the bars specify the mutation counts.