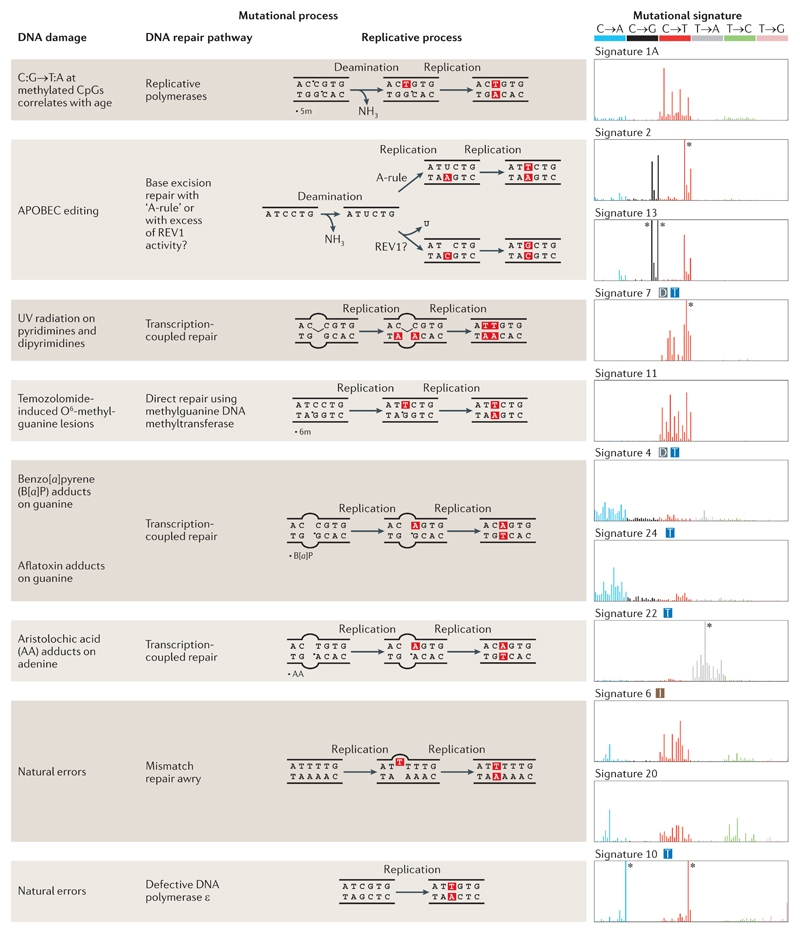
Figure 2. Summary of known mutational signatures, and the components of DNA damage and repair that constitute the mutational processes.
There are marked differences among the 96-element mutational signatures, which are dominated by specific elements, including enrichment of various base substitutions (shown in the graphs on the right), transcriptional strand bias (T), excess of dinucleotide mutations (D), and association with insertions and deletions (I). The asterisks mark instances at which the limits of the y axes, which represent the likelihood of specific mutations being present in a signature, are exceeded. 5m, 5′ methyl group; 6m, O6 methyl group; APOBEC, apolipoprotein B mRNA editing enzyme, catalytic polypeptide; REV1, DNA repair protein REV1; UV, ultraviolet.