Figure 5. Gene rearrangements in cancer.
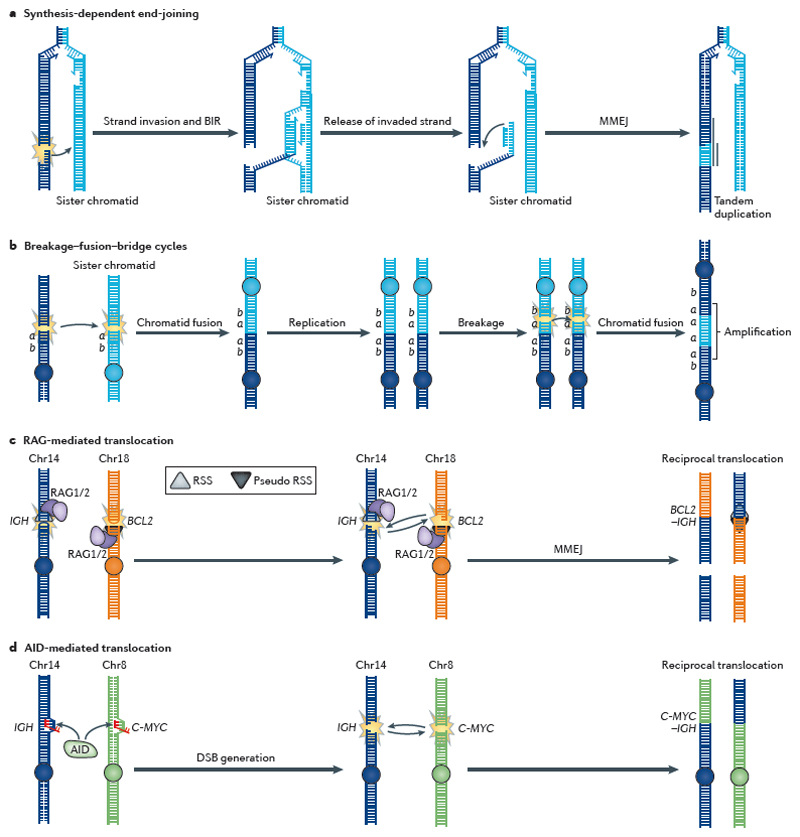
Gene rearrangements in cancer arise primarily from DNA double-strand breaks (DSBs). a | Synthesis-dependent end-joining (SDEJ), which is involved in repairing replication-associated DSBs, results in tandem duplications. Break-induced replication (BIR) initiates synthesis on the sister chromatid after strand invasion. Reversed branch migration of the Holliday junction formed following strand invasion can release the invaded strand, which contains extra DNA material from the sister chromatid and is fused to the original end by microhomology-mediated end-joining (MMEJ), resulting in a tandem duplication90. b | Sister chromatid fusion causes gene amplification by breakage–fusion–bridge cycles96,97. In this process, two adjacent DSBs on sister chromatids are substrates for non-homologous end-joining (NHEJ), which rejoins the sister chromatids. After replication, these are again broken to form anther fusion chromosome carrying four gene a copies. c | The V(D)J recombination-activating (RAG) proteins recognize either the correct recombination signal sequences (RSSs) or almost identical (that is, pseudo) RSSs at which they initiate DSBs; they then mediate interchromosomal translocation rather than regular recombination within the V(D)J segments. This can create an immunoglobulin H (IGH)–BCL2 (B-cell CLL/lymphoma 2) fusion gene that drives cancer. d | Activation-induced cytidine deaminase (AID) is involved in class switch recombination and deaminates cytosines to uracils in transcribed regions, which are then processed by DNA repair enzymes into a DSB. If DSBs coexist in the IGH and C-MYC genes, then they can recombine by interchromosomal translocation to produce an IGH–C-MYC fusion gene. Chr, chromosome.