Abstract
Efficient enrichment glycoproteins/glycopeptides from complex biological solutions are very important in the biomedical sciences, in particular biomarker research. In this work, the high hydrophilic polyethylenimine conjugated polymaltose polymer brushes functionalized magnetic Fe3O4 nanoparticles (NPs) denoted as Fe3O4–PEI–pMaltose were designed and synthesized via a simple two-step modification. The obtained superhydrophilic Fe3O4–PEI–pMaltose NPs displayed outstanding advantages in the enrichment of N-linked glycopeptides, including high selectivity (1:100, mass ratios of HRP and bovine serum albumin (BSA) digest), low detection limit (10 fmol), large binding capacity (200 mg/g), and high enrichment recovery (above 85%). The above-mentioned excellent performance of novel Fe3O4–PEI–pMaltose NPs was attributed to graft of maltose polymer brushes and efficient assembly strategy. Moreover, Fe3O4–PEI–pMaltose NPs were further utilized to selectively enrich glycopeptides from human renal mesangial cell (HRMC, 200 μg) tryptic digest, and 449 N-linked glycopeptides, representing 323 different glycoproteins and 476 glycosylation sites, were identified. It was expected that the as-synthesized Fe3O4–PEI–pMaltose NPs, possessing excellent performance (high binding capacity, good selectivity, low detection limit, high enrichment recovery, and easy magnetic separation) coupled to a facile preparation procedure, have a huge potential in N-glycosylation proteome analysis of complex biological samples.
Introduction
Protein N-glycosylation, as one of the most common and significant post-translational modifications, plays an important role in biological processes, such as cell signal transduction, protein folding, cell recognition, etc.1−3 Aberrant protein N-glycosylation is frequently involved in many major human diseases, including cancer, Alzheimer’s disease (AD), and infectious disease.4,5 Therefore, the efficient isolation and identification of N-glycopeptides is especially beneficial for understating their biological functions and for the discovery of new clinical biomarkers and therapeutic drug targets. Currently, mass spectrometry (MS) is a powerful and effective tool in proteomics which provides the possibility to analyze the N-glycoproteome.6−8 However, owing to the matrix complexity of biological samples, a low abundance of glycoproteins, and severe ion signal suppression of nonglycopeptides, it remains still an analytical challenge to comprehensively characterize glycoproteins. Therefore, an effective enrichment of glycopepetides prior to MS analysis becomes imperative to elucidate the structures of glycans and clarify glycan-attached sites.
The common enrichment strategies based on glycan-specific recognition or glycan physicochemical properties for glycosylated proteins/peptides, including lectin affinity,9−12 hydrazide chemistry,13−15 boronic acid chemistry,16−21 and hydrophilic interaction liquid chromatography (HILIC),22−25 have been developed. Among them, HILIC has aroused much attention for glycopeptides enrichment by utilizing the strong hydrophilicity of the glycopeptides and HILIC materials, due to its broad glycan specificity, excellent reproducibility, and good MS compatibility.26,27 Until now, a number of HILIC nanomaterials have been synthesized by introducing hydrophilic functional groups onto the surface of mesoporous silica, graphene oxide, metal–organic frameworks, and magnetic nanoparticles.28−35 In virtue of their strong magnetic responsibility, good biocompatibility, easy and versatile modification, Fe3O4 nanoparticles (NPs) based on magnetic separation has become an effective isolation technique in proteomic research.36−38 The hydrophilic ligands, immobilized on magnetic nanoparticles, would simultaneously achieve fast separation and low loss of N-linked glycopeptides from a complex sample under an external magnetic field. However, most of HILIC adsorbents need tedious synthesis steps and harsh conditions to acquire the functional moieties; this leads to relative low binding capacity and enrichment selectivity. It has been reported that more hydrophilic functional groups grafted on the surface of HILIC substrates lead to a better performance of glycopeptides from the highly complex biosamples.23 Therefore, there is great demand to obtain ultrahydrophilic nanocomposites with more functional groups by a facile synthesis procedure for specific enrichment, especially for N-linked glycopeptide enrichment in complex samples.
Herein, a new type of maltose-functionalized hydrophilic magnetic nanoparticles, Fe3O4–polyethylenimine–polymaltose denoted as Fe3O4–PEI–pMaltose, was assembled by a facile strategy (Scheme 1). Briefly, PEI–coated magnetic Fe3O4 NPs were prepared by solvothermal reaction, then succinic anhydride was reacted with the surface amino groups of PEI. Maltose polymer brushes (Scheme S1, Supporting Information) were grafted on the surface of magnetic Fe3O4 NPs via an esterification reaction. The abundant maltose on the surface of Fe3O4 NPs could specifically enrich glycopeptides, and the magnetic core makes the NPs separate easily from solution under an external magnetic field. In addition, the hydrophilic polymer can provide low adsorption of nonglycopeptides, which ensures the novel nanocomposite with high selectivity, sensitivity, large binding capacity, and high recovery for N-glycopeptides enrichment.
Scheme 1. Schematic Illustration of the Fabrication of Fe3O4–PEI–pMaltose NPs and the Selective Enrichment Process for the N-Linked Glycopeptides.
Experimental Section
Materials
Horseradish peroxidase (HRP), immunoglobulin G (IgG), peptide-N4-(N-acetyl-β-d-glucosaminyl) asparagine amidase F (PNGase F) and bovine serum albumin (BSA) were obtained from Sigma-Aldrich, USA. Polyethylenemine (PEI, Mw = 70 000) was obtained from Alfa Aesar, Tianjin, China. Dithiothreitol (DTT), urea, ammonium bicarbonate (NH4HCO3), and iodoacetamide (IAA) were purchased from Solarbio, China. Trifluoroacetic acid (TFA), Amberlite IR 120 and 2-bromoethanol were purchased from J&K, China. N,N,N′,N″,N″-Pentamethyldiethylenetriamine (PMDETA) and 2-bromo-2-methylpropionyl bromide were from Aladdin, China. 2,5-Dihydroxybenzoic acid (DHB) was purchased from TCI, Japan. Trypsin was from Sangon Biotech Co. Led., China. Dicyclohexylcarbodiimide (DCC) and N-hydroxysuccinimide (NHS) were obtained from Shanghai Medpep. Co., China. Iron(III) chloride hexahydrate (FeCl3·6H2O), sodium sulfate (Na2SO4), succinic anhydride, tetrahydrofuran (THF), triethylamine (Et3N), dimethyl sulfoxide (DMSO), d-(+)-maltose and other analytical grade reagents were obtained from Tianjin Chemical Reagent Factory, China. Deionized water (18.25 MΩ cm) was purified with a Milli-Q water system, Millipore, Milford, MA, USA.
Characterization
Transmission electron microscope (TEM) characterization was carried out on a JEOL JEM-2100 EX transmission electron microscope (Japan). Scanning electron microscope (SEM) measurement was performed on a JSM-6360LV scanning electron microscope (Japan). Hydrodynamic diameter (Dh) measurement was performed with a Brookhaven BI-200SM instrument (USA). Fourier transform infrared (FT-IR) spectra in KBr were recorded using the BRUKER TENSOR 27 Fourier transform infrared spectrophotometer. The crystal structure of nanoparticles was determined on a Rigaku D/max/2500v/pc, Japan. The X-ray diffraction (XRD) pattern was carried out on a Rigaku D/max/2500 X-ray diffractometer, Japan. The X-ray photoelectron spectra (XPS) was obtained on a Shimadzu Kratos AXIS Ultra DLD X-ray photoelectron spectrometer, Japan. The magnetic properties were analyzed with a LDJ9600-1 vibrating sample magnetometer, USA. The hydrophilicity was evaluated with JCY-1 contact angle analyzer, China. Zeta potential was measured by a Brookhaven ZetaPALS potentiometric analyzer, USA.
MALDI-TOF MS measurements were performed on a Bruker AutoflexIII LRF200-CID instrument, Germany. DHB (25 mg/mL, V(ACN)/V(H2O)/V(TFA) = 80:19:1) was used as matrix. All the LC–MS/MS analyses were carried out on a Orbitrap Q-Exactive mass spectrometer, Thermo Fisher Scientific, Waltham, MA.
Preparation of Fe3O4–PEI NPs
Fe3O4–PEI NPs were synthesized by the solvothermal method.34 Typically, 1.8 g of FeCl3·6H2O, 7.2 g of NaAc, and 1.8 g of PEI were dissolved in 90 mL of EG under sonication to give a homogeneous solution which was then stirred mechanically for 30 min at 60 °C. Then the resulting solution was poured into a autoclave and reacted at 220 °C for 2 h. Deionized water alternates with ethanol three times to wash the Fe3O4–PEI NPs. Finally, the resulting Fe3O4–PEI NPs were dried in vacuum oven.
Preparation of Fe3O4–PEI–COOH NPs
The succinic anhydride (2.5 g) was added into the solution of Fe3O4–PEI NPs (500 mg) in dry THF (120 mL). The dispersion solution was refluxed for 12 h. The obtained Fe3O4–PEI–COOH NPs were washed several times using deionized water and ethanol and dried in vacuum oven.
Synthesis of Fe3O4–PEI–pMaltose NPs
The succinic anhydride (500 mg) was added into the THF solution (22 mL) containing Fe3O4–PEI–COOH NPs (100 mg). After ultrasonic dispersion uniformity, the solution was reacted at 60 °C for 12 h. The activated Fe3O4–PEI–COOH NPs were washed with dry THF and were dispersed in dry THF (120 mL) containing maltose polymer (Supporting Information, 200 mg) and Et3N (800 μL). After reaction at 60 °C for 4 h, the Fe3O4–PEI–pMaltose NPs were obtained.
Digestion of Proteins
Standard proteins (HRP, IgG, and BSA) were digested according to the previous literature.33 Briefly, the proteins were dissolved in 50 mM of NH4HCO3 solution and denatured by boiling or 8 M urea. The denatured proteins were reduced with DTT and alkylated by IAA in sequence. After emzymolysis with trypsin, the peptide solution was frozen at −20 °C for standby application.
One mg protein extraction from human renal mesangial cells (HRMC) was precipitated by TCA. The pellet was resuspended in 100 mM of NH4HCO3. The proteins underwent reduction, alkylation, and emzymolysis in sequence. The resulting digests were desalted and enriched using Sep-pak C18 cartridges (Waters Ltd., Elstree, UK), evaporated to dryness.
Isotope Dimethylation Labeling of Human IgG Tryptic Digest
Stable isotope dimethyl labeling was performed as the following procedures: After the C18 StageTip desalting step, tryptic peptides of IgG (100 μg) were dissolved in sodium acetate buffer (100 μL, 100 mM, pH 5–6), and then transferred in equal amount into two tubes. The solutions (50 μL) were mixed with 8 μL of CH2O (4%, v/v) or 8 μL of CD2O (4%, v/v), which were labeled as light (L) and heavy (H), separately. After a brief vortexing, 8 μL of NaBH3CN solution (0.6 M) in water was added to the L and H labeled samples, respectively. The reactions were terminated with 8 μL of 4% ammonia solution after shaking for 1 h.
Glycopeptides Enrichment under Hydrophilic Mode
Fe3O4–PEI–pMaltose NPs (or Fe3O4–PEI, Fe3O4–PEI–COOH NPs, 15 μg) was washed thrice with loading buffer (V(ACN)/V(H2O)/V(TFA) = 92:7.9:0.1) and dispersed in the above buffer (400 μL) containing a determined amount of standard protein digests. After being incubated for 30 min, the NPs were washed thrice with the 400 μL loading buffer. Finally, the N-glycopeptides captured by Fe3O4–PEI–pMaltose NPs were released by elution buffer (2 × 13 μL, V(ACN)/V(H2O)/V(TFA) = 30:69.9:0.1) for 6 min. The supernatant was collected, lyophilized, redissolved in 4 μL of elution buffer and analyzed by MALDI-TOF MS.
For the glycopeptide enrichment from human renal mesangial cells, 200 μg of the digests was dissolved in 6 mL loading buffer (80% ACN/H2O, 0.1% FA), incubated with 20 mg of Fe3O4–PEI–pMaltose NPs for 1 h, and subsequently washed thrice with 2 mL of loading buffer. Then, the trapped glycopeptides were eluted twice with 400 μL of elution buffer for 30 min, and the elution was evaporated to dryness. The obtained glycopeptides were redissolved in 10 mM NH4HCO3, and glycan moieties were removed by 1000 units of PNGase F. The mixture was desalted and enriched using Sep-pak C18 cartridges (Waters Ltd., Elstree, UK), evaporated to dryness, and redissolved prior to analysis by nano LC–MS/MS.
Results and Discussion
Preparation and Characterization of Fe3O4–PEI–pMaltose NPs
The size and morphology of obtained Fe3O4–PEI, Fe3O4–PEI–COOH and Fe3O4–PEI–pMaltose NPs were characterized by TEM and SEM. As shown in Figure 1, after the modification of short chain carboxylic acid and maltose polymer brush on the layer of Fe3O4–PEI NPs with covalent bond, the corresponding NPs still exhibited good dispersity and almost no changes in particle sizes. Also, from the TEM image, the diameter of the resulting Fe3O4–PEI–pMaltose NPs was about 60–100 nm, which is consistent with the hydrodynamic diameter by dynamic light scattering technique (Figure S1). Because the maltose polymer was prepared by atom transfer radical polymerization (ATRP) in the presence of a mole ratio of 20:1 for maltose monomer and initiator bis(2-bromoisobutyryl) hexanediamide (Supporting Information), the polymer lines of pMaltose are short, which did not result in great size change of Fe3O4–PEI–pMaltose NPs after immobilization on the surface of magnetic Fe3O4–PEI NPs.
Figure 1.
TEM imagesof Fe3O4–PEI (a), Fe3O4–PEI–COOH (b), Fe3O4–PEI–pMaltose (c), NPs and SEM image of Fe3O4–PEI (d), Fe3O4–PEI–COOH (e), Fe3O4–PEI–pMaltose (f) NPs, respectively.
The zeta potentials of Fe3O4–PEI, Fe3O4–PEI–COOH and Fe3O4–PEI–pMaltose NPs were monitored to further demonstrate successful modification of Fe3O4 NPs. The zeta potentials of Fe3O4–PEI, Fe3O4–PEI–COOH and Fe3O4–PEI–pMaltose NPs were −25.54 ±1.23, −33.51, ±1.31, and −19.00 ± 0.86 mV in alkaline solution, respectively (Figure S2, Supporting Information). PEI and maltose polymer are cationic polyelectrolytes, and carboxylic acid is an anionic electrolyte. Because of the cationic PEI and maltose polymer, magnetic nanoparticles (Fe3O4–PEI, Fe3O4–PEI–COOH) had smaller negative potential than Fe3O4–PEI–COOH modified with anionic carboxyl groups in alkaline condition (pH 10.5).
To further confirm the functionalization of magnetic Fe3O4 NPs, Fourier transform infrared (FT-IR) spectrometry was performed for Fe3O4–PEI, Fe3O4–PEI–COOH, and Fe3O4–PEI–pMaltose NPs, respectively (Figure 2). In the spectrum of Figure 2a, the peaks at 585 and 440 cm–1 were ascribed to the stretching vibration Fe–O bond (Fe3+ bond and Fe2+ bond, respectively), which was consistent with frequency bands of the spinel ferrite phase of Fe3O4 while the Fe–O band for γ-Fe2O3 is usually seen at 540 cm–1.39 The broad peak centered at 3428 cm–1 was assigned to the stretching vibration of N–H and/or OH bonds. The peaks at 1622 and 1557 cm–1 were attributed to the stretching vibration of the C=O bond and deformation vibration of N–H of amide group, respectively. Compared with the above spectrum of Fe3O4–PEI, the new broad peak at 3448–2964 cm–1 of the carboxyl group and the enhanced peak at 1557 cm–1 of the amide group in the spectrum of Fe3O4–PEI–COOH NPs (Figure 2b) revealed the successful modification of carboxyl groups on the surface of Fe3O4–PEI. Meanwhile, the disappearance of carboxyl groups (Figure 2c) indicated that the maltose polymer brushes were grafted on the surface of the nanoparticles.
Figure 2.

FT-IR spectra of Fe3O4–PEI (a), Fe3O4–PEI–COOH (b) and Fe3O4–PEI–pMaltose (c) NPs.
The magnetic hysteresis curves (Figure 3) indicated that the nanomaterials (Fe3O4–PEI, Fe3O4–PEI–COOH, and Fe3O4–PEI–pMaltose) possessed nearly superparamagnetic properties with saturation magnetization (Ms) of 48.25, 46.53, and 45.3 emu·g–1. The Fe3O4–PEI–pMaltose NPs can be easily separated within 10 s with an external magnetic field (Figure 3 inset) and quickly redispersed after removal of the magnetic field.
Figure 3.
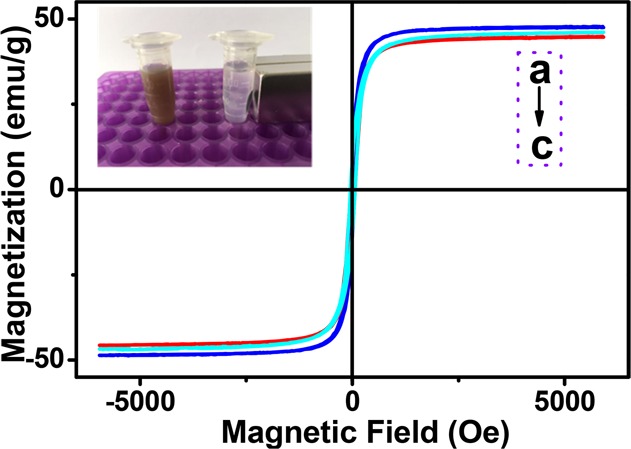
Magnetic hysteresis curves of Fe3O4–PEI (a), Fe3O4–PEI–COOH (b), and Fe3O4–PEI–pMaltose (c) NPs.
Identification of the crystalline phases of Fe3O4–PEI, Fe3O4–PEI–COOH and Fe3O4–PEI–pMaltose NPs was performed by wide-angle X-ray diffraction (XRD) analysis (Figure S3, Supporting Information). The characteristic diffraction peaks of Fe3O4 (2θ = 30.0°, 35.4°, 43.1°, 53.4°, 57.0°, 62.6°) at the corresponding 2θ values were indexed as (220), (311), (400), (422), (511), and (440), respectively, which were in accordance with that of standard magnetite XRD pattern (JCPDS card, file No. 19-0629). Although the XRD patterns of the products clearly exhibit the spinel structure, it is difficult to distinguish the crystalline structure between Fe3O4 and the γ-Fe2O3 phase only from the XRD patterns because of their similarity.40 The XPS technique was applied to character the magnetic products because XPS is very sensitive to Fe2+ and Fe3+ ions. The data of XPS (Figure 4a) of Fe3O4–PEI NPs showed a C12 peak around 282.7 eV, O 1s peak at 530.6 eV, N 1s peak at 398.5 eV, and Fe signals at about 56.6 eV for Fe 3p, 710.9 and 724.5 eV for Fe 2p3/2 and Fe 2p1/2 states, respectively (Figure 4b). The absence of the satellite peaks on the magnifying pattern of Fe (2p) also confirmed the formation of Fe3O4 rather than γ-Fe2O3,40−42 which was consistent with the results from the FT-IR spectra.
Figure 4.
XPS spectrum (a) and high-resolution Fe (2p) binding energy spectrum (b) of Fe3O4–PEI NPs.
The thermogravimetric analysis (TGA) curves of Fe3O4–PEI, Fe3O4–PEI–COOH, Fe3O4–PEI–pMaltose NPs are shown in Figure 5. It can be seen that 4.4% weight loss occurred for Fe3O4–PEI NPs (blue curve) corresponding to the content of PEI onto the nanoparticles surface. And there was 9.5 and 13.4% weight loss for Fe3O4–PEI–COOH, Fe3O4–PEI–pMaltose, respectively. From the data of TGA curves, the amount of maltose grafted onto the Fe3O4–PEI–pMaltose NPs was calculated to 108.2 μmol·g–1.
Figure 5.

TGA curves of Fe3O4–PEI (a), Fe3O4–PEI–COOH (b) and Fe3O4–PEI–pMaltose (c) NPs.
Hydrophilicity of nanoparticles is a key evaluation factor for enrichment performance. Herein, the contact angle of prepared Fe3O4–PEI, Fe3O4–PEI–pMaltose NPs were measured with the powder tabletting method. The contact angle of Fe3O4–PEI and Fe3O4–PEI–pMaltose NPs were 42.6° and 14.7°, respectively (Figure 6). As expected, the angle distinctly decreased after modification, indicating that Fe3O4 NPs functionalized polymer maltose brush has superior hydrophilicity which could enhance the enrichment effect.
Figure 6.

Water contact angle of Fe3O4–PEI (a) and Fe3O4–PEI–pMaltose (b) NPs.
Glycopeptide Enrichment from Standard Proteins by Fe3O4–PEI–pMaltose NPs
To manifest enrichment based on the hydrophilic interaction between the hydrophilicity of polymer maltose brush and targets, three synthetic nanoparticles (Fe3O4–PEI, Fe3O4–PEI–COOH, Fe3O4–PEI–pMaltose NPs) were used to enrich the glycopeptides from the standard HRP tryptic digest under the same conditions. Figure 7a shows the direct analysis of HRP digest (100 fmol) without the enrichment procedure. The signal peaks of low abundance of glycopeptides were completely suppressed, and no target analyte was detected. After enrichment Fe3O4–PEI, Fe3O4–PEI–COOH, Fe3O4–PEI–pMaltose NPs, and three and five glycopeptides were distinctly identified and most abundant nonglycopeptides were efficiently removed (Figure 7b–e). Detailed information on glycopeptides enriched by Fe3O4–PEI–pMaltose NPs from HRP tryptic digest is displayed in Table S1 (Supporting Information). The greater number and enhanced signal intensity of glycopeptides were observed for Fe3O4–PEI–pMaltose NPs in comparsion with those for Fe3O4–PEI and Fe3O4–PEI–COOH. After the HRP tryptic digest was adsorbed by Fe3O4–PEI–pMaltose NPs, no glycopeptide peaks were found in the residual solution (Figure 7c). The results demonstrated the good hydrophilicity Fe3O4–PEI–pMaltose NPs has as highly specific to glycopeptides.
Figure 7.

MALDI-TOF mass analysis of tryptic digest of HRP (100 fmol): (a) before enrichment, (b) eluent, and (c) supernatant after enrichment with Fe3O4–PEI–pMaltose NPs; eluent after enrichment with (d) Fe3O4–PEI, and (e) Fe3O4–PEI–COOH NPs. The peaks of glycopeptides are marked with a red inverted triangle.
The detection limit of Fe3O4–PEI–pMaltose NPs for N-glycopeptides enrichment was investigated with different concentrations of HRP tryptic digest. As shown in Figure 8a, five target glycopeptides with high signal intensity were identified when the concentration of HRP digests was 50 fmol. Even when the concentration of HRP digests was as low as 10 fmol, five target glycopeptides were still observed (Figure 8b).
Figure 8.

MALDI-TOF mass analysis of different concentrations of HRP tryptic digests: (a) 50, (b) 10 fmol HRP after enrichment with Fe3O4–PEI–pMaltose NPs. The peaks of glycopeptides are marked with a red inverted triangle.
To further evaluate the enrichment selectivity of Fe3O4–PEI–pMaltose NPs between nonglycopeptides and glycopeptides, the mixture of HRP and BSA tryptic digest with different mass ratios was investigated. As shown in Figure 9, seven glycopeptides were identified from tryptic BSA and HRP at ratios of 1:1 and 10:1 after enrichment by Fe3O4–PEI–pMaltose NPs. When the ratios were increased to 50:1 and 100:1, five glycopeptides and four glycopeptides were still detected, respectively, albeit with trace nonglycopeptide signals (Figure 9c,d). The results demonstrated that the Fe3O4–PEI–pMaltose NPs has great potential for N-glycopeptides enrichment from complex biological samples.
Figure 9.

MALDI-TOF mass analysis of tryptic BSA and HRP after enrichment. The mass ratios of BSA/HRP are 1:1 (a), 10:1 (b), 50:1 (c), and 100:1 (d). The peaks of glycopeptides are marked with a red inverted triangle.
To demonstrated that Fe3O4–PEI–pMaltose NPs have no bias toward different kinds of glycans, tryptic IgG (20 pmol) which contains a different glycoform from HRP was employed. Before enrichment, merely two glycopeptides were detected with low peak intensities, and there existed a great number of interferences from nonglycopeptides (Figure 10a). Seventeen high signal-to-noise glycopeptides were obtained after enrichment by Fe3O4–PEI–pMaltose NPs (Figure 10b, detailed information is listed in Table S2, Supporting Information).The eluted glycopeptides were deglycosylated by PNGaseF, and two strong signals of deglycosylated peptides were detected (Figure 10c).
Figure 10.

MALDI-TOF mass analysis of tryptic digests of IgG (20pmol): direct analysis (a), after enrichment with Fe3O4–PEI–pMaltose NPs (b), and deglycosylation by PNGase F (c). The peaks of glycopeptides are marked with a blue inverted triangle.
Evaluation of Binding Capacity of Fe3O4-DA-Maltose NPs for Glycopeptide
Different amounts (5–30 μg) of Fe3O4–PEI–pMaltose NPs were used to treat 3 μg of IgG digest. The elution was analyzed by MALDI-TOF MS. When the peak intensity of representative glycopeptides reached maximum, the total amount of glycopeptides were bonded onto the NPs. The binding capacity was calculated by 3 μg IgG digest to NPs. As shown in Figure 11, the N-glycopeptides from 3 μg IgG digest were captured by 15 μg of Fe3O4–PEI–pMaltose NPs, therefore the binding capacity of Fe3O4–PEI–pMaltose NPs for N-glycopeptides was about 200 mg g–1, which greatly exceeded the binding capacity of other maltose functionalized Fe3O4 materials such as Fe3O4@SiO2@PEG-Maltose,23−26 Fe3O4-DA-Maltose,33 and Fe3O4-PEI-Maltose,34 NPs.
Figure 11.

Intensity of six selected N-glycopeptides from tryptic digests of human IgG (3 μg) after enrichment by different amount of Fe3O4–PEI–pMaltose NPs.
The Reusability and Stability of Fe3O4–PEI–pMaltose NPs
To evaluate the reusability and stability, Fe3O4–PEI–pMaltose NPs placed for two month at room temperature were applied to enrich standard HRP glycopeptides from tryptic digest in consecutive times. As shown in Figure S4 (Supporting Information), the N-linked glycopeptides were clearly detected in the first time and sixth time run, indicating that as-prepared Fe3O4–PEI–pMaltose NPs owned excellent repeatability and long-term stability for N-linked glycopeptides enrichment.
Enrichment Recovery of Glycopeptides by Fe3O4–PEI–pMaltose NPs
The stable-isotope dimethyl labeling samples were used to study the enrichment recovery of Fe3O4–PEI–pMaltose NPs for glycopeptides. Briefly, the heavy-tagged tryptic digest was enriched with Fe3O4–PEI–pMaltose NPs, and then the eluted N-glycopeptides were mixed up with the light-tagged tryptic digest. The mixed dimethyl labeling samples were enriched with Fe3O4–PEI–pMaltose NPs, followed with elution, deglycosylation by PNGaseF, and MALDI-TOF analysis. The recovery yield (H/L) was calculated by the peak intensity ratio of heavy isotope-labeled peptides to the corresponding light isotope-labeled peptides. Meanwhile, the recovery yield (L/H) was also evaluated (Figure S5, Table S3, Supporting Information). The recovery yields (L/H or H/L) of two deglycosylated stable-isotope dimethyl labeling peptides (m/z = 1186.0, 1218.0 or 1190.1, 1222.1) were about 89% or 85%, respectively, which confirmed that Fe3O4–PEI–pMaltose NPs have great potential for N-glycopeptides enrichment.
Glycopeptide Enrichment from Human Renal Mesangial Cell (HRMC) Tryptic Digest by Fe3O4–PEI–pMaltose NPs
Human renal mesangial cells serve as a filtration barrier of the kidney. The injury of mesangial cells could cause diabetic nephropathy, leading end-stage renal disease. Emerging evidence indicates that mesangial cells can be damaged by high glucose, however the mechanism is unclear. Given such outstanding performance in standard glycopeptides enrichment, Fe3O4–PEI–pMaltose NPs were applied to enrich glycopeptides from human renal mesangial cell tryptic digest, treated with high glucose, and followed by LC–MS/MS analysis (cell culture, protein extraction, MS/MS data analysis, see Supporting Information). Followed by the Uniprot-Human protein sequence database for analysis, 449 N-linked glycopeptides, representing 323 different glycoproteins and 476 glycosylation sites were identified. The number of proteins and glycosylation sites detected extremely exceeded those of our previous Fe3O4-DA-Maltose NPs.33 Detailed information on glycan structures of human renal mesangial cells after enrichment by Fe3O4–PEI–pMaltose NPs is listed in Table S4 (Supporting Information).
Conclusions
In summary, novel polymer maltose brushes-interspersed Fe3O4 magnetic nanoparticles (Fe3O4–PEI–pMaltose NPs) were synthesized successfully via a facile two-step method for selective enrichment of N-glycopeptides. On the basis of its good biocompatibility, excellent hydrophilicity, and magnetic responsibility, Fe3O4–PEI–pMaltose functionalized with a hyperbranched PEI structure and high maltose polymer chains loading exhibited distinctly improved selectivity, sensitivity, large binding capacity, and good recovery for HRP and IgG digests. A total of 449 N-linked glycopeptides, representing 323 different glycoproteins and 476 glycosylation sites were found from human renal mesangial cell tryptic digest. It can be expected that the Fe3O4–PEI–pMaltose NPs would hold great potential in N-glycoproteome research.
Acknowledgments
We gratefully appreciate the financial support by the National Natural Science Foundation of China (No 21475067), the Natural Science Foundation of Tianjin (No. 15JCYBJC20600), and the CAMS Innovation Fund for Medical Sciences (CIFMS, 2017-I2M-3-019 and 2016-I2M-3-022).
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsomega.7b01788.
Detailed procedure of cell culture and protein extraction, LC–MS/MS analysis and MS/MS data analysis; schematic illustration of maltose polymer brush with ATRP polymerization and synthesis of maltose polymer brush; hydrodynamic diameter histograms of Fe3O4–PEI–pMaltose NPs; MALDI-TOF MS spectra of the isotope dimethylation labeled human IgG peptides after deglycosylation by PNGaseF; zeta potentials, XRD patterns of Fe3O4–PEI, Fe3O4–PEI–COOH and Fe3O4–PEI–pMaltose NPs; the repeatability performance of the Fe3O4–PEI–pMaltose NPs for enrichemnt of 100 fmol tryptic HRP in consecutive times; the recovery of N-glycosylation motifs from tryptic digests of IgG enriched by Fe3O4–PEI–pMaltose NPs; identified glycopeptides and glycan structures of human HRP, IgG digests and human renal mesangial cells after enrichment by Fe3O4–PEI–pMaltose NPs (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Roth J.; Zuber C.; Park S.; Jang I.; Lee Y.; Kysela K. G.; Le Fourn V.; Santimaria R.; Guhl B.; Cho J. W. Protein N-Glycosylation, Protein Folding, and Protein Quality Control. Mol. Cells 2010, 30, 497–506. 10.1007/s10059-010-0159-z. [DOI] [PubMed] [Google Scholar]
- Demetriou M.; Granovsky M.; Quaggin S.; Dennis J. W. Negative Regulation of T-cell Activation and Autoimmunity by Mgat5 N-Glycosylation. Nature 2001, 409, 733–739. 10.1038/35055582. [DOI] [PubMed] [Google Scholar]
- Partidge E. A.; Roy C. L.; Guglielmo G. M.; Pawling J.; Cheung P.; Granovsky M.; Nabi I. R.; Wrana J. L.; Dennis J. W. Regulation of Cytokine Receptors by Golgi N-Glycan Processing and Endocytosis. Science 2004, 306, 120–124. 10.1126/science.1102109. [DOI] [PubMed] [Google Scholar]
- Ohtsubo K.; Marth J. D. Glycosylation in Cellular Mechanisms of Health and Disease. Cell 2006, 5, 855–867. 10.1016/j.cell.2006.08.019. [DOI] [PubMed] [Google Scholar]
- Nita-Lazar M.; Noonan V.; Rebustini I.; Walker J.; Menko A. S.; Kukuruzinska M. A. Overexpression of DPAGT1 Leads to Aberrant N-Glycosylation of E-Cadherin and Cellular Discohesion in Oral Cancer. Cancer Res. 2009, 14, 5673–5680. 10.1158/0008-5472.CAN-08-4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dell A.; Morris H. R. Glycoprotein Structure Determination by Mass Spectrometry. Science 2001, 291, 2351–2356. 10.1126/science.1058890. [DOI] [PubMed] [Google Scholar]
- Baycin-Hizal D.; Tian Y.; Akan I.; Jacobson E.; Clark D.; Wu A.; Jampol R.; Palter K.; Betenbaugh M.; Zhang H. Glycofish: A Database of Zebrafish N-linked Glycoproteins Identified Using SPEG Method Coupled with LC/MS. Anal. Chem. 2011, 83, 5296–5303. 10.1021/ac200726q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding W.; Hill J. J.; Kelly J. Selective Enrichment of Glycopeptides from Glycoprotein Digests Using Ion-Pairing Normal-Phase Liquid Chromatography. Anal. Chem. 2007, 79, 8891–8899. 10.1021/ac0707535. [DOI] [PubMed] [Google Scholar]
- Dela Rosa M. A.; Chen W. C.; Chen Y. J.; Obena R. P.; Chang C. H.; Capangpangan R. Y.; Su T. H.; Chen C. L.; Chen P. J.; Chen Y. J. One-Pot Two-Nanoprobe Assay Uncovers Targeted Glycoprotein Biosignature. Anal. Chem. 2017, 89, 3973–3980. 10.1021/acs.analchem.6b04396. [DOI] [PubMed] [Google Scholar]
- Kaji H.; Yamauchi Y.; Takahashi N.; Isobe T. Mass Spectrometric Identification of N-Linked Glycopeptides Using Lectin-Mediated Affinity Capture and Glycosylation Site-Specific Stable Isotope Tagging. Nat. Protoc. 2006, 6, 3019–3027. 10.1038/nprot.2006.444. [DOI] [PubMed] [Google Scholar]
- Lu Y. W.; Chien C. W.; Lin P. C.; Huang L. D.; Chen C. Y.; Wu S. W.; Han C. L.; Khoo K. H.; Lin C. C.; Chen Y. J. Bad-Lectins: Boronic Acid-Decorated Lectins with Enhanced Binding Affinity for the Selective Enrichment of Glycoproteins. Anal. Chem. 2013, 85, 8268–8276. 10.1021/ac401581u. [DOI] [PubMed] [Google Scholar]
- Kim J. Y.; Kim S. K.; Kang D.; Moon M. H. Dual Lectin-Based Size Sorting Strategy to Enrich Targeted N-Glycopeptides by Asymmetrical Flow Field-Flow Fractionation:Profiling Lung Cancer Biomarkers. Anal. Chem. 2012, 84, 5343–5350. 10.1021/ac300772w. [DOI] [PubMed] [Google Scholar]
- Huang J. F.; Wan H.; Yao Y. T.; Li J. N.; Cheng K.; Mao J. W.; Chen J.; Wang Y.; Qin H. Q.; Zhang W. B.; Ye M. L.; Zou H. F. Highly Efficient Release of Glycopeptides from Hydrazide Beads by Hydroxylamine Assisted PNGase F Deglycosylation for N-Glycoproteome Analysis. Anal. Chem. 2015, 87, 10199–10204. 10.1021/acs.analchem.5b02669. [DOI] [PubMed] [Google Scholar]
- Liu L. T.; Yu M.; Zhang Y.; Wang C. C.; Lu H. J. Hydrazide Functionalized Core-Shell Magnetic Nanocomposites for Highly Specific Enrichment of N-Glycopeptides. ACS Appl. Mater. Interfaces 2014, 6, 7823–7832. 10.1021/am501110e. [DOI] [PubMed] [Google Scholar]
- Zhang L. J.; Jiang H. C.; Yao J.; Wang Y. L.; Fang C. Y.; Yang P. Y.; Lu H. J. Highly Specific Enrichment of N-Linked Glycopeptides Based on Hydrazide Functionalized Soluble Nanopolymers. Chem. Commun. 2014, 50, 1027–1029. 10.1039/C3CC47347C. [DOI] [PubMed] [Google Scholar]
- Zhang X. H.; Wang J. W.; He X. W.; Chen L. X.; Zhang Y. K. Tailor-Made Boronic Acid Functionalized Magnetic Nanoparticles with a Tunable Polymer Shell-Assisted for the Selective Enrichment of Glycoproteins/Glycopeptides. ACS Appl. Mater. Interfaces 2015, 7, 24576–24584. 10.1021/acsami.5b06445. [DOI] [PubMed] [Google Scholar]
- Wang Y. L.; Liu M. B.; Xie L. Q.; Fang C. Y.; Xiong H. M.; Lu H. J. Highly Efficient Enrichment Method for Glycopeptide Analyses: Using Specific and Nonspecific Nanoparticles Synergistically. Anal. Chem. 2014, 86, 2057–2064. 10.1021/ac403236q. [DOI] [PubMed] [Google Scholar]
- Ma R. N.; Hu J. J.; Cai Z. W.; Ju H. X. Facile Synthesis of Boronic Acid-Functionalized Magnetic Carbon Nanotubes for Highly Specific Enrichment of Glycopeptides. Nanoscale 2014, 6, 3150–3156. 10.1039/c3nr05367a. [DOI] [PubMed] [Google Scholar]
- Wang J. X.; Wang Y. N.; Gao M. X.; Zhang X. M.; Yang P. Y. Multilayer Hydrophilic Poly(phenol-formaldehyde resin)-Coated Magnetic Graphene for Boronic Acid Immobilization as aNovel Matrix for Glycoproteome Analysis. ACS Appl. Mater. Interfaces 2015, 7, 16011–16017. 10.1021/acsami.5b04295. [DOI] [PubMed] [Google Scholar]
- Lin Z. A.; Pang J. L.; Yang H. H.; Cai Z. W.; Zhang L.; Chen G. N. One-Pot Synthesis of an Organic-Inorganic Hybrid Affinity Monolithic Column for Specific Capture of Glycoproteins. Chem. Commun. 2011, 47, 9675–9677. 10.1039/c1cc13082j. [DOI] [PubMed] [Google Scholar]
- Lin Z. A.; Wang J.; Tan X. Q.; Sun L. X.; Yu R. F.; Yang H. H.; Chen G. N. Preparation of Boronate-Functionalized Molecularly Imprinted Monolithic Column with Polydopamine Coating for Glycoprotein Recognition and Enrichment. J. Chromatogr. A 2013, 1319, 141–147. 10.1016/j.chroma.2013.10.059. [DOI] [PubMed] [Google Scholar]
- Yu L.; Li X.; Guo Z.; Zhang X.; Liang X. Hydrophilic Interaction Chromatography Based Enrichment of Glycopeptides by Using Click Maltose: A Matrix with High Selectivity and Glycosylation Heterogeneity Coverage. Chem. - Eur. J. 2009, 15, 12618–12626. 10.1002/chem.200902370. [DOI] [PubMed] [Google Scholar]
- Xiong Z. C.; Zhao L.; Wang F. J.; Zhu J.; Qin H. Q.; Wu R. A.; Zhang W. B.; Zou H. F. Synthesis of Branched PEG Brushes Hybrid Hydrophilic Magnetic Nanoparticles for the Selective Enrichment of N-linked Glycopeptides. Chem. Commun. 2012, 48, 8138–8140. 10.1039/c2cc33600f. [DOI] [PubMed] [Google Scholar]
- Chen R.; Seebun D.; Ye M. L.; Zou H. F.; Figeys D. Site-Specific Characterization of Cell Membrane N-Glycosylation with Integrated Hydrophilic Interaction Chromatography Solid Phase Extraction and LC-MS/MS. J. Proteomics 2014, 103, 194–203. 10.1016/j.jprot.2014.03.040. [DOI] [PubMed] [Google Scholar]
- Selman M. H. J.; Hemayatkar M.; Deelder A. M.; Wuhrer M. Cotton HILIC SPE Microtips for Microscale Purification and Enrichment of Glycans and Glycopeptides. Anal. Chem. 2011, 83, 2492–2499. 10.1021/ac1027116. [DOI] [PubMed] [Google Scholar]
- Ma W.; Xu L. N.; Li Z.; Sun Y. L.; Bai Y.; Liu H. W. Post-Synthetic Modification of an Amino-Functionalized Metal-Organic Framework for Highly Efficient Enrichment of N-Linked Glycopeptides. Nanoscale 2016, 8, 10908–10912. 10.1039/C6NR02490D. [DOI] [PubMed] [Google Scholar]
- Jin T.; Xiong Z. C.; Zhu X.; Mehio N.; Chen Y. J.; Hu J.; Zhang W. B.; Zou H. F.; Liu H. L.; Dai S. Template-Free Synthesis of Mesoporous Polymers for Highly Selective Enrichment of Glycopeptides. ACS Macro Lett. 2015, 5, 570–574. 10.1021/acsmacrolett.5b00235. [DOI] [PubMed] [Google Scholar]
- Wan H.; Huang J. F.; Liu Z. S.; Li J. N.; Zhang W. B.; Zou H. F. A Dendrimer-Assisted Magnetic Graphene-Silica Hydrophilic Composite for Efficient and Selective Enrichment of Glycopeptides from the Complex Sample. Chem. Commun. 2015, 51, 9391–9394. 10.1039/C5CC01980J. [DOI] [PubMed] [Google Scholar]
- Ma W.; Xu L. N.; Li X. J.; Shen S. S.; Wu M.; Bai Y.; Liu H. W. Cysteine-Functionalized Metal-Organic Framework: Facile Synthesis and High Efficient Enrichment of N-linked Glycopeptides in Cell Lysate. ACS Appl. Mater. Interfaces 2017, 9, 19562–19568. 10.1021/acsami.7b02853. [DOI] [PubMed] [Google Scholar]
- Sun N. R.; Wang J. W.; Yao J. Z.; Deng C. H. Hydrophilic Mesoporous Silica Materials for Highly Specific Enrichment of N-Linked Glycopeptide. Anal. Chem. 2017, 89, 1764–1771. 10.1021/acs.analchem.6b04054. [DOI] [PubMed] [Google Scholar]
- Jiang B.; Liang Y.; Wu Q.; Jiang H.; Yang K. G.; Zhang L. H.; Liang Z.; Peng X. J.; Zhang Y. K. New GO-PEI–Au-L-Cys ZIC-HILIC Composites: Synthesis and Selective Enrichment of Glycopeptides. Nanoscale 2014, 6, 5616–5619. 10.1039/C4NR00274A. [DOI] [PubMed] [Google Scholar]
- Li J. N.; Wang F. J.; Liu J.; Xiong Z. C.; Huang G.; Wan H.; Liu Z. Y.; Cheng K.; Zou H. F. Functionalizing with glycopeptide dendrimers significantly enhances the hydrophilicity of the magnetic nanoparticles. Chem. Commun. 2015, 51, 4093–4096. 10.1039/C5CC00187K. [DOI] [PubMed] [Google Scholar]
- Bi C. F.; Zhao Y. R.; Shen L. J.; Zhang K.; He X. W.; Chen L. X.; Zhang Y. K. Click Synthesis of Hydrophilic Maltose-Functionalized Iron Oxide Magnetic Nanoparticles Based on Dopamine Anchors for Highly Selective Enrichment of Glycopeptides. ACS Appl. Mater. Interfaces 2015, 7, 24670–24678. 10.1021/acsami.5b06991. [DOI] [PubMed] [Google Scholar]
- Li J. N.; Wang F. J.; Wan H. J.; Liu J.; Liu Z. Y.; Cheng K.; Zou H. F. Magnetic Nanoparticles Coated with Maltose-Functionalized Polyethyleneimine for Highly Efficient Enrichment of N-Glycopeptides. J. Chromatogr. A 2015, 1425, 213–220. 10.1016/j.chroma.2015.11.044. [DOI] [PubMed] [Google Scholar]
- Yeh C. H.; Chen S. H.; Li D. T.; Lin H. P.; Huang H. J.; Chang C. I.; Shih W. L.; Chern C. L.; Shi F. K.; Hsu J. L. Magnetic Bead-Based Hydrophilic Interaction Liquid Chromatography of Glycopeptide Enrichments. J. Chromatogr. A 2012, 1224, 70–78. 10.1016/j.chroma.2011.12.057. [DOI] [PubMed] [Google Scholar]
- Li Y.; Zhang X. M.; Deng C. H. Functionalized Magnetic Nanoparticles for Sample Preparation in Proteomics and Peptidomics Analysis. Chem. Soc. Rev. 2013, 21, 8517–8539. 10.1039/c3cs60156k. [DOI] [PubMed] [Google Scholar]
- Lin Z. A.; Zheng J. N.; Lin F.; Zhang L.; Cai Z. W.; Chen G. N. Synthesis of Magnetic Nanoparticles with Immobilized Aminophenylboronic Acid for Selective Capture of Glycoproteins. J. Mater. Chem. 2011, 21, 518–524. 10.1039/C0JM02300K. [DOI] [Google Scholar]
- Gao C. H.; Lin G.; Lei Z. X.; Zheng Q.; Lin J. S.; Lin Z. A. Facile Synthesis of Core-Shell Structured Magnetic Covalent Organic Framework Composite Nanospheres for Selective Enrichment of Peptides with Simultaneous Exclusion of Proteins. J. Mater. Chem. B 2017, 5, 7496–7503. 10.1039/C7TB01807J. [DOI] [PubMed] [Google Scholar]
- Mohapatra J.; Mitra A.; Bahadur D.; Aslam M. Surface Controlled Synthesis of MFe2O4 (M = Mn, Fe, Co, Ni and Zn) Nanoparticles and Their Magnetic Characteristics. CrystEngComm 2013, 15, 524–532. 10.1039/C2CE25957E. [DOI] [Google Scholar]
- Lu J.; Jiao X. L.; Chen D. R.; Li W. Solvothermal Synthesis and Characterization of Fe3O4 and γ-Fe2O3 Nanoplates. J. Phys. Chem. C 2009, 113, 4012–4017. 10.1021/jp810583e. [DOI] [Google Scholar]
- Luo J. S.; Liu J. L.; Zeng Z. Y.; Ng C. F.; Ma L. J.; Zhang H.; Lin J. Y.; Shen Z. X.; Fan H. J. Three-Dimensional Graphene Foam Supported Fe3O4 Lithium Battery Anodes with Long Cycle Life and High Rate Capability. Nano Lett. 2013, 13, 6136–6143. 10.1021/nl403461n. [DOI] [PubMed] [Google Scholar]
- Zhang M.; Chen L. F.; Zheng J.; Li W. Z.; Hayat T.; Alharbi N. S.; Gan W. J.; Xu J. L. The Fabrication and Application of Magnetite Coated N-Doped Carbon Microtubes Hybrid Nanomaterials with Sandwich Structures. Dalton Trans. 2017, 46, 9172–9179. 10.1039/C7DT01155E. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.






