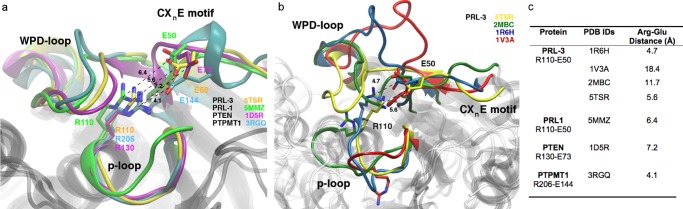
Figure 1.
(a) Structural superposition of PTPs and the CXnE motif. The respective glutamic acids and the catalytic arginine residues are indicated for PRL-3 (orange: 5TSR), PRL-1 (green: 5MMZ), PTEN (magenta: 1D5R), and PTPMT1 (cyan: 3RGQ). Tight glutamate–arginine interactions as seen with PTPMT1 seem rather questionable for PRL-3, PRL-1, and PTEN. For PTPMT1, Glu144 from the “EEYE” motif aligns well with the Glu of the CXnE motif of the other PTPs. (b) Superposition of an X-ray (5TSR) and various NMR structures (others) of PRL-3 showing the protein’s flexibility for active-site conformations. The respective position of Arg110 and Glu50 is indicated. (c) Distances between Arg and Glu residues of the indicated phosphatases. For 5TSR, the reported value is an average calculated on both chains A and C; for 2MBC and 1R6H, the reported value is the average value calculated on the deposited 20 models.