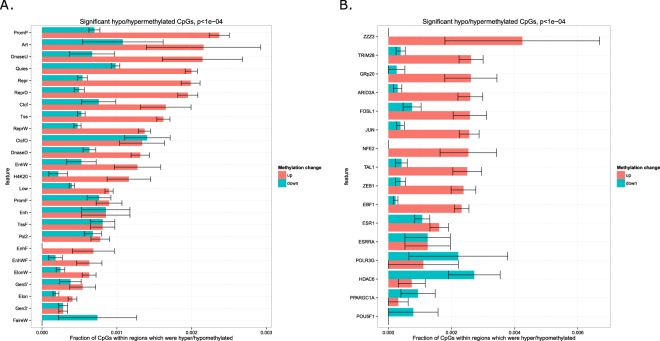
Figure 4.
Distribution of hypo- and hypermethylated CpGs within certain epigenomic features. Fraction of significantly hypo- (blue bars) and hypermethylated (red bars) CpGs among all CpGs within a given set of genomic regions. (A) Regions of ChromHMM genome segmentation. (B). Transcription factor binding sites (only the sites with the highest hyper- and hypomethylation rates are plotted). CpGs that were significantly hypermethylated after VHL inactivation, were enriched in AP-1 (JUN/FOS) and TRIM28 binding sites. Hypomethylated CpGs were enriched in HDAC6 binding sites.