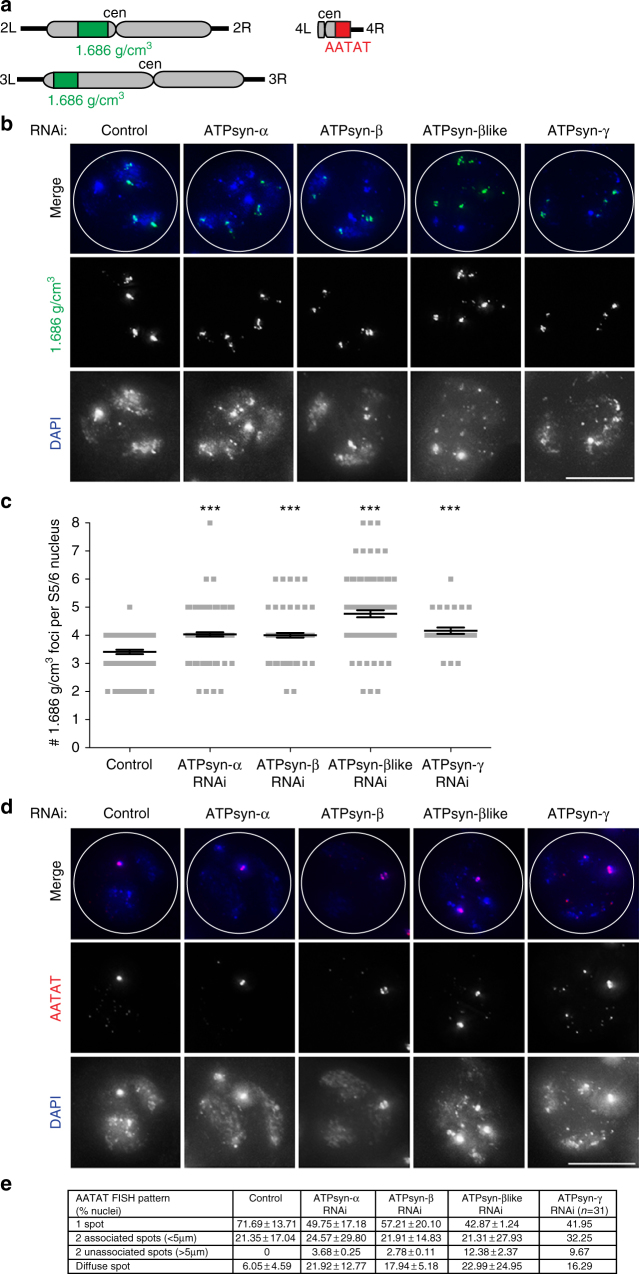
Fig. 3.
Arm cohesion defects upon ATP synthase-α/-β/-βlike/-γ RNAi. a Cartoon showing chromosomal location of FISH probes used in the study. The 1.686 g/cm3 satellite probe (green) targets both the second (2 L) and third (3 L) chromosome arms. The AATAT repeat probe (red) targets the fourth (4 R) chromosome arm. b Micrograph of 1.686 g/cm3 FISH probe (green) performed on control S5/6 nuclei or nuclei RNAi-depleted at 25 °C of ATPsyn-α, -β, -βlike and –γ. DNA is stained with DAPI; white circle outlines the nucleus (n = 3). Scale bar = 10 μm. c Quantitation of 1.686 g/cm3 foci in control S5/6 nuclei (n = 81) or nuclei RNAi-depleted for ATPsyn-α (n = 122), -β (n = 95), -βlike (n = 102) and –γ (n = 31). Data pooled from two individual experiments. Significance was determined using an unpaired Student's t-test, ***p < 0.001. Error bars = SEM. d Micrograph of AATAT FISH probe (red) performed on control S5/6 nuclei or nuclei RNAi-depleted of ATPsyn-α, -β, -βlike and -γ. DNA is stained with DAPI; white circle outlines the nucleus (n = 3). Scale bar = 10 μm. e Quantitation (% nuclei ± SD) of the AATAT hybridisation pattern in control S5/6 nuclei or nuclei RNAi-depleted of ATPsyn-α, -β, -βlike (n = 2, 50 nuclei quantified per experiment) and -γ (n = 31 nuclei)