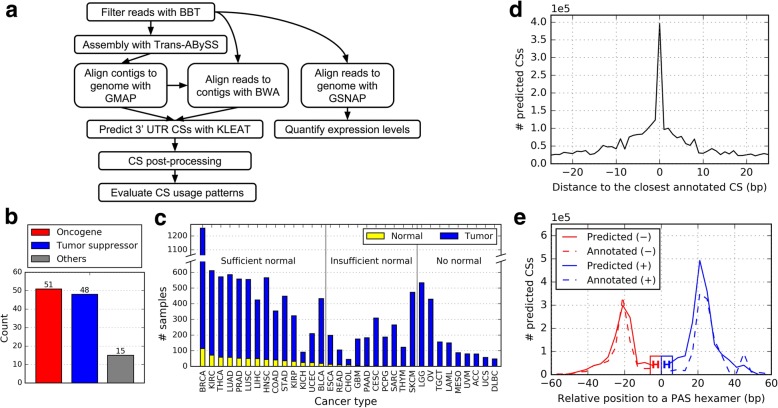
Fig. 1.
Cleavage site predictions. (a) Schematic diagram of the CS prediction pipeline. See Additional file 1: Figure S3A for a description of the CS post-processing step. (b) Count of gene types. (c) Count of TCGA RNA-Seq samples across 33 cancer types (sorted in decreasing order of normal and tumor samples). Sufficient normal: ≥15 samples. Alphabetically, ACC: adrenocortical carcinoma; BLCA: bladder urothelial carcinoma; BRCA: breast invasive carcinoma; CESC: cervical squamous cell carcinoma and endocervical adenocarcinoma; CHOL: cholangiocarcinoma; COAD: colon adenocarcinoma; DLBC: lymphoid neoplasm diffuse large B-cell lymphoma; ESCA: esophageal carcinoma; GBM: glioblastoma multiforme; HNSC: head and neck squamous cell carcinoma; KICH: kidney chromophobe; KIRC: kidney renal clear cell carcinoma; KIRP: kidney renal papillary cell carcinoma; LAML: acute myeloid leukemia; LGG: brain lower grade glioma; LIHC: liver hepatocellular carcinoma; LUAD: lung adenocarcinoma; LUSC: lung squamous cell carcinoma; MESO: mesothelioma; OV: ovarian serous cystadenocarcinoma; PAAD: pancreatic adenocarcinoma; PCPG: pheochromocytoma and paraganglioma; PRAD: prostate adenocarcinoma; READ: rectum adenocarcinoma; SARC: sarcoma; SKCM: skin cutaneous melanoma; STAD: stomach adenocarcinoma; TGCT: testicular germ cell tumors; THCA: thyroid carcinoma; THYM: thymoma; UCEC: uterine corpus endometrioid carcinoma; UCS: uterine carcinosarcoma; UVM: uveal melanoma. (d, e) Validation of our pipeline for predicting CSs. (d) Distribution of the distances between predicted and the closest annotated CSs. (e) Distribution of the distances between a predicted CS and the PAS hexamer motif found within 50 bp upstream. A high-resolution version of this figure is available for download in Additional file 5