Figure 4. Diverse structural rearrangements of the AR axis.
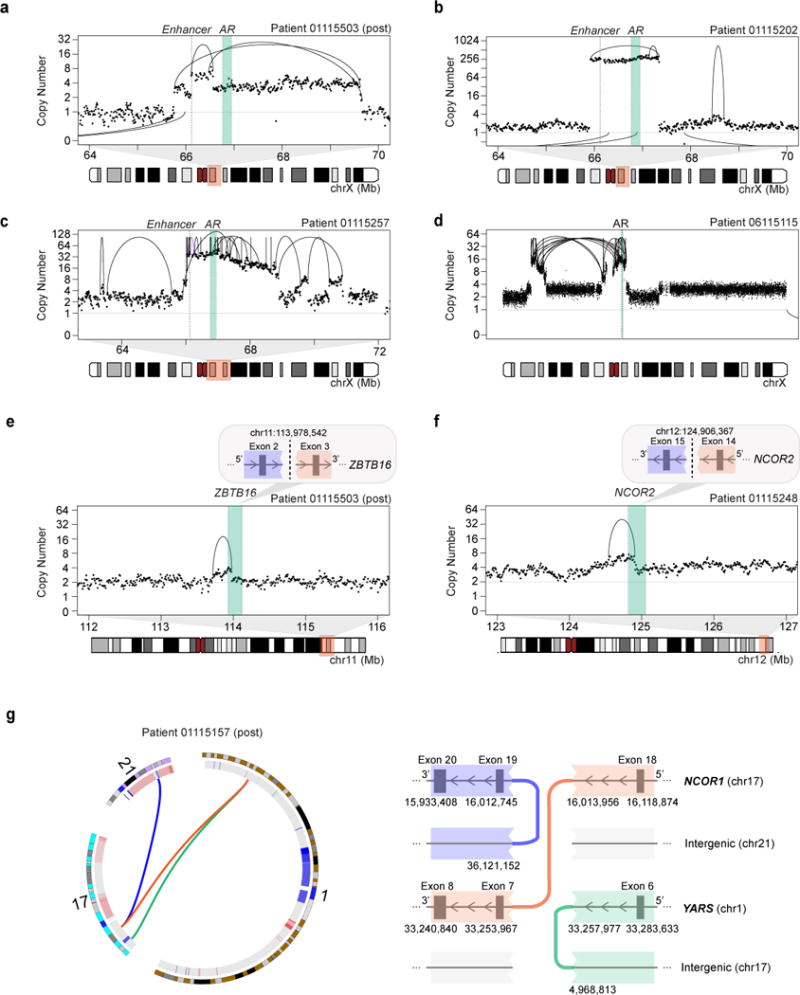
(a–d) Rearrangements involving the AR locus include: (a) simple and nested duplications, (b) high-level copy number gains, (c) amplification due to breakage-fusion-bridge cycles, and (d) trans-centromeric rearrangements. Copy number shown is purity-adjusted.
(e–g) Examples of rearrangements disrupting AR-related genes in mCRPC include duplications transecting (e) ZBTB16 and (f) NCOR2, and (g) a chained chromoplexy event resulting in disruption of the C-terminal domain of NCOR1 and production of an in-frame N-terminal NCOR1-YARS fusion transcript. Inter-chromosomal rearrangements are shown as arcs below the data points. Note: Samples 01115503, 01115202, 01115257, and 01115248 all display the TDP.
See also Figure S5 and Table S5.
