FIGURE 1.
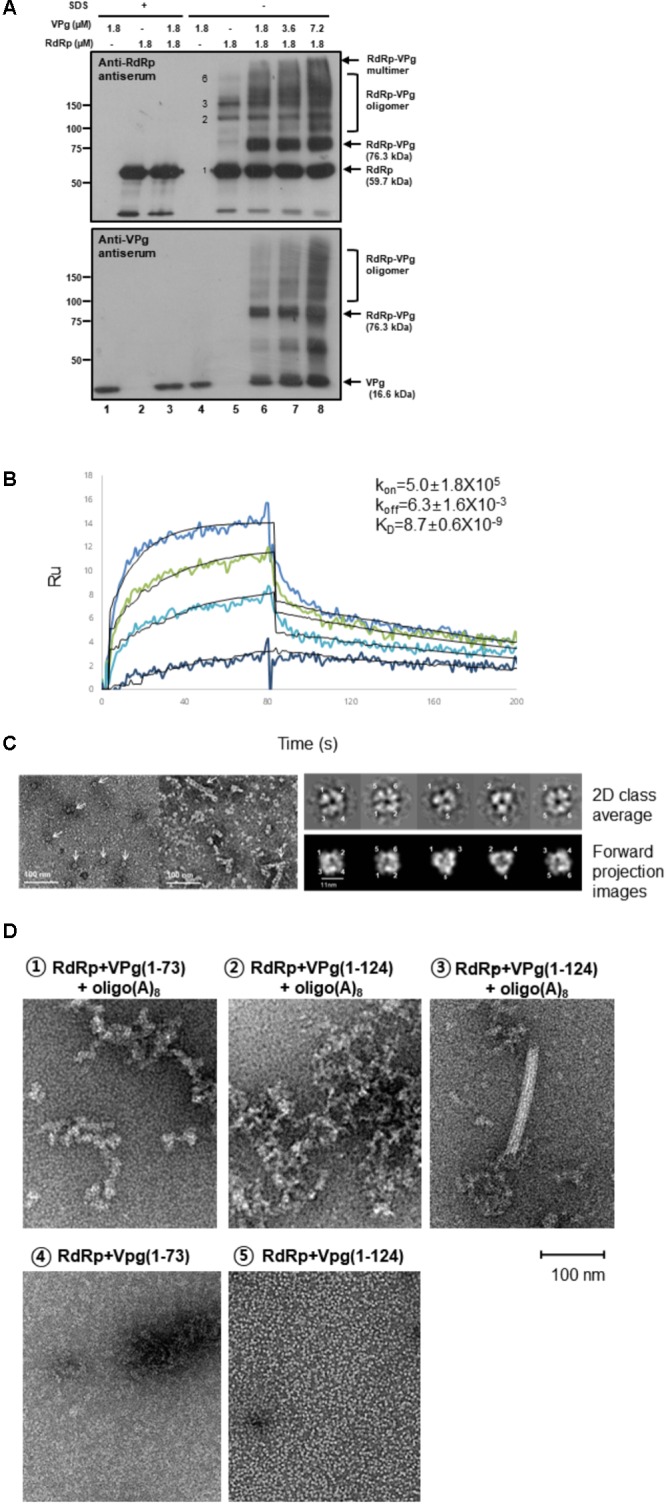
Interactions in the RdRp-VPg(1-73) complex. (A) Cross-linking studies of RdRp-VPg-RNA complexes. The mixture containing 50 mM HEPES (pH 7.4), 5 mM MgCl2, 5 mM DTT, 1 μM oligo(A)15, 2 μM RdRp, and 6 μM VPg was incubated with or without 0.5% SDS. The reaction mixture was cross-linked by 0.001% glutaraldehyde treatment at room temperature for 6 min. Proteins were separated on 8% SDS-PAGE and analyzed by western blotting with anti-RdRp or anti-VPg antisera. The molecular masses of standard proteins are shown in kDa unit on the left. The RdRp, VPg monomer, and RdRp-VPg complex bands are indicated on the right of the gel. Monomer, dimer, trimer, and hexamer bands of RdRp were indicated with numbers 1, 2, 3, and 6, respectively. The experiments were repeated in triplicates. (B) SPR binding assay. RdRp or mutant proteins in sodium acetate buffer (pH 5.0) were immobilized onto a CM5 chip after being serially diluted in 20 mM Tris–HCl, pH 8.0, 100 mM NaCl, and 5 mM MgCl2. The binding affinity constant (KD) was determined from the association (kon) and dissociation (koff) rates by evaluating the 1:1 Langmuir binding model kinetics in the sensorgrams. Colored curves depict experimental data at different analyte concentrations and fitted curves modeled to describe a 1:1 binding event are overlaid in black. The experiments were duplicated starting from two separate protein preparations and each measurement performed in triplicate. (C) Electron microscopic studies of RdRp-VPg-RNA complexes. (Left panel) Hexamers of MNV RdRp are shown on the left (pointed by arrows) and hexameric oligomers are shown on the right (arrows). 1–6 μM proteins were used and the reaction mixture was incubated for 1 h at room temperature, and the reaction mixture was placed on carbon-coated copper grids. Images were collected with a 4K × 4K Eagle HS CCD camera on a Tecnai T120 microscope (FEI, Eindhoven, Netherlands) operating at 120 kV. The scale bar represents 100 nm. (Right panel) Single particles of RdRp hexamers are represented as 2D class averages and (lower panel) corresponding forward projection images. The six monomers of RdRp are numbered from 1 to 6 based on crystal structures in Figure 2D. (D) Electron microscopic images of the RdRp-VPg(1-73)-oligo(A)8 or RdRp-VPg(1-124)-oligo(A)8 complexes are shown (1D, 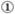 and
and 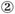 ). An RdRp tubular fibril (approximately 18 nm in diameter) formed after incubation with VPg(1-124) and oligo(A)8 is shown (1D,
). An RdRp tubular fibril (approximately 18 nm in diameter) formed after incubation with VPg(1-124) and oligo(A)8 is shown (1D,  ). A comparison between VPg(1-73) and VPg(1-124) in complex with RdRp is shown in the absence of oligo(A)8 (1D,
). A comparison between VPg(1-73) and VPg(1-124) in complex with RdRp is shown in the absence of oligo(A)8 (1D,  and
and 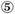 ).
).
