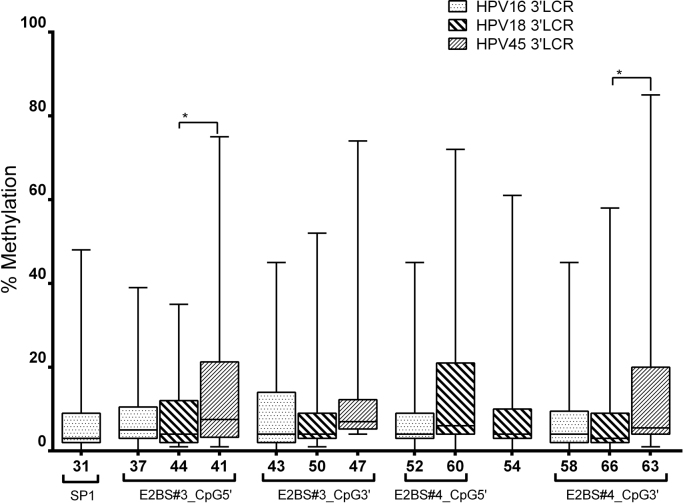
Fig. S1.
Comparison of the methylation between homologous CpG sites at the 3′LCR of HPV in disrupted samples. A higher methylation in E2BS#3 and E2BS#4 of HPV45 (nt 41 and 63) was observed than in HPV18 (nt 44 and 66, respectively); p = 0.0163 and p = 0.0246, respectively. Homologous CpG sites in E2 binding site motifs of each HPV genotype comprised: (i) nt 37, 44 and 41 of HPV16, HPV18 and HPV45, respectively, at the 5′ end of E2BS#3; (ii) nt 43, 50 and 47 of HPV16, HPV18 and HPV45, respectively, at the 3′ end of E2BS#3; (iii) nt 52 and 60 of HPV16 and HPV18, respectively, at the 5′ end of E2BS#4; and (iv) nt 58, 66 and 63 of HPV16, HPV18 and HPV45, respectively, at the 3′ end of E2BS#4. Associations approaching borderline statistical significance: nt 47 vs 43 (p = 0.0668), nt 60 vs 52 (p = 0.0995) and 63 vs 58 (p = 0.0716).